Beyond the Petri Dish: Innovative Strategies for Cultivating the Unculturable Microbial Majority
This article provides a comprehensive overview of the latest scientific advances and methodologies for cultivating previously unculturable microorganisms.
Beyond the Petri Dish: Innovative Strategies for Cultivating the Unculturable Microbial Majority
Abstract
This article provides a comprehensive overview of the latest scientific advances and methodologies for cultivating previously unculturable microorganisms. Aimed at researchers, scientists, and drug development professionals, it explores the foundational reasons for unculturability, details cutting-edge cultivation techniques like high-throughput dilution-to-extinction and in-situ encapsulation, and offers optimization strategies for microbial growth. It further addresses the critical validation of cultivation success through genomic comparisons and highlights the profound implications for discovering novel antibiotics, such as clovibactin, and for expanding our functional understanding of microbial ecology and evolution.
The Uncultured Majority: Understanding the Scale and Significance of Hidden Microbial Life
For researchers in microbiology and drug development, a fundamental challenge persists: the vast majority of microbial diversity on Earth resists growth under standard laboratory conditions. This phenomenon, known as the "Great Plate Count Anomaly," describes the discrepancy of several orders of magnitude between the number of microbial cells observed microscopically in an environmental sample and the number of colonies that actually grow on a Petri plate [1] [2]. It is estimated that while traditional cultivation methods allow us to study only about 0.1% to 1.0% of microbial species in many environments, the remaining 99% represent an untapped reservoir of genetic and metabolic diversity, including potential novel natural products [1] [3]. This technical support center is designed to provide actionable troubleshooting guides and methodologies to help you overcome these cultivation barriers and access this "uncultured majority."
FAQs: Understanding the Core Challenge
1. What exactly does "unculturable" mean in a practical research context?
"Unculturable" is a operational term indicating that a microorganism cannot be grown using current standard laboratory culturing techniques. It does not mean the organism can never be cultured [1]. The term highlights a gap in our knowledge about the organism's specific biological and environmental needs. These microbes are often metabolically active in their native environment but fail to proliferate when transferred to artificial media [4].
2. What are the primary reasons our laboratory cultivation attempts fail?
Failure is typically not due to a single factor but a combination of several:
- Inappropriate Nutrient Levels: Standard laboratory media are often too nutrient-rich, which can inhibit or kill oligotrophic (nutrient-preferring) bacteria from environments like open ocean or soil [5] [2].
- Absence of Essential Growth Factors: The laboratory medium may lack specific signaling molecules, vitamins, or other growth factors that are provided by other organisms in a complex community [1] [4].
- Dependence on Other Microorganisms (Syntrophy): Some bacteria rely on the metabolic byproducts of helper species for growth. Isolating them disrupts these essential cross-feeding relationships [2] [4].
- Viable But Non-Culturable (VBNC) State: Many bacteria enter a dormant, low-metabolism state in response to stress and will not form colonies on plates, even though they are alive [3].
3. How can we identify if an uncultured microbe is even present in our sample?
Cultivation-independent molecular techniques are key for detecting these organisms.
- 16S rRNA Gene Sequencing: This is the primary tool. By extracting DNA directly from an environmental sample (e.g., soil, water), amplifying the 16S rRNA gene, and sequencing it, you can identify the phylogenetic "fingerprints" of microbes without needing to culture them [1] [6].
- Metagenomics: Shotgun sequencing of all the DNA in a sample allows you to reconstruct full or partial genomes of uncultured organisms, providing hypotheses about their metabolic capabilities and potential growth requirements [7] [6].
4. Why is overcoming this anomaly critical for drug discovery?
Bacteria are a prolific source of bioactive natural products. The "discovery void" of new antibiotic classes since the late 1980s is partly attributed to the repeated isolation of the same, culturable bacteria [1]. Uncultured phyla are believed to harbor the majority of microbial natural product diversity, and accessing them is essential for discovering new classes of antibiotics, anticancer agents, and other pharmaceuticals [1].
Troubleshooting Guides & Experimental Protocols
Guide 1: Low Microbial Yield on Standard Media
Problem: The number of colonies on your plates is far lower than the direct cell count from your sample.
Solutions:
- Reduce Nutrient Concentration: Use diluted standard media (e.g., 1/10 or 1/100 strength R2A) or create low-nutrient media using filtered water from your sample's environment [5] [2].
- Prolong Incubation Time: Many slow-growing bacteria will not form visible colonies in the standard 1-2 day incubation period. Extend incubation times to several weeks or even months [4].
- Use Alternative Gelling Agents: Agar can contain inhibitory compounds. Test gellan gum as a solidifying agent, which has been shown to improve the growth of some fastidious microbes [4].
- Supplement with Soil or Environmental Extract: Add filter-sterilized extract from the sample's native environment (e.g., soil extract) to the medium to introduce natural nutrients and trace elements [4].
Guide 2: Cultivating a Specific, Previously Uncultured Phylotype
Problem: Molecular data (e.g., 16S sequencing) confirms the presence of a target uncultured organism (e.g., from the TM7 phylum or SAR11 clade), but you cannot get it to grow.
Solutions:
- Employ Co-culture Techniques: Cultivate your sample in the presence of a "helper" strain from the same environment. The helper strain may provide essential growth factors, remove toxic metabolites, or signal resuscitation from dormancy [1] [8].
- Simulate the Natural Environment with Diffusion Chambers/Bioreactors: Trap cells in a semi-permeable chamber that is then placed back into the native environment or an artificial system that mimics it. This allows diffusion of natural nutrients and signals while containing the cells for later isolation [1] [4].
- Apply High-Throughput Culturing (HTC) in Microplates: Use extinction culturing by serially diluting cells to a very low density (e.g., average of 1-5 cells per well) in 48- or 96-well plates containing a low-nutrient medium. This separates cells and reduces competition, allowing slow-growers to proliferate [5].
Detailed Experimental Protocols
Objective: To isolate oligotrophic and slow-growing bacteria from aquatic environments.
Materials:
- Filter-sterilized (0.2 µm) water from the sample environment, used as the cultivation medium.
- 48-well or 96-well non-tissue-culture-treated microplates.
- DAPI (4',6-diamidino-2-phenylindole) stain and fluorescence microscope.
Method:
- Prepare Medium: Collect environmental water, filter-sterilize it, and restore bicarbonate buffer by sparging with COâ‚‚ followed by sterile air.
- Enumerate Inoculum: Perform a direct cell count of the source sample using DAPI staining and microscopy.
- Dilute and Dispense: Dilute the sample in the prepared medium to a final average inoculum of 1-5 cells per well. Dispense 1 mL aliquots into each well of the microplate.
- Incubate: Incubate plates in the dark at a temperature relevant to the sample's environment (e.g., 16°C for temperate marine waters) for 3-6 weeks.
- Detect Growth: After incubation, use a cell array manifold or plate reader to detect growth. For the array method, filter 200 µL from each well onto a polycarbonate membrane, stain with DAPI, and score for microcolonies under fluorescence microscopy.
- Isolate and Purify: Transfer cells from positive wells to fresh medium or solid media for further purification and identification.
Objective: To cultivate soil bacteria by maintaining a continuous connection to their natural chemical environment.
Materials:
- Inner chamber (e.g., 2L plastic container) with multiple 6mm holes drilled in its walls.
- Outer chamber (e.g., 4L plastic container).
- Polycarbonate membrane (0.4 µm pore size).
- Fresh, sieved soil from the sample site.
- Low-nutrient cultivation media (e.g., R2A, soil extract media).
Method:
- Assemble Bioreactor: Glue the polycarbonate membrane to the outer surface of the inner chamber, covering all holes. Sterilize all components.
- Load Chambers: Place the inner chamber inside the outer chamber. Fill the gap between the two chambers with the fresh, sieved soil.
- Inoculate: Add your soil sample and cultivation medium into the inner chamber.
- Incubate: Seal the bioreactor and incubate at room temperature with gentle stirring for 4 weeks. During this time, chemical compounds, nutrients, and signaling molecules diffuse from the soil through the membrane into the inner chamber.
- Sample and Isolate: After incubation, take samples from the liquid in the inner chamber, perform serial dilutions, and plate them onto solid agar plates. Incubate these plates for several weeks to isolate pure cultures.
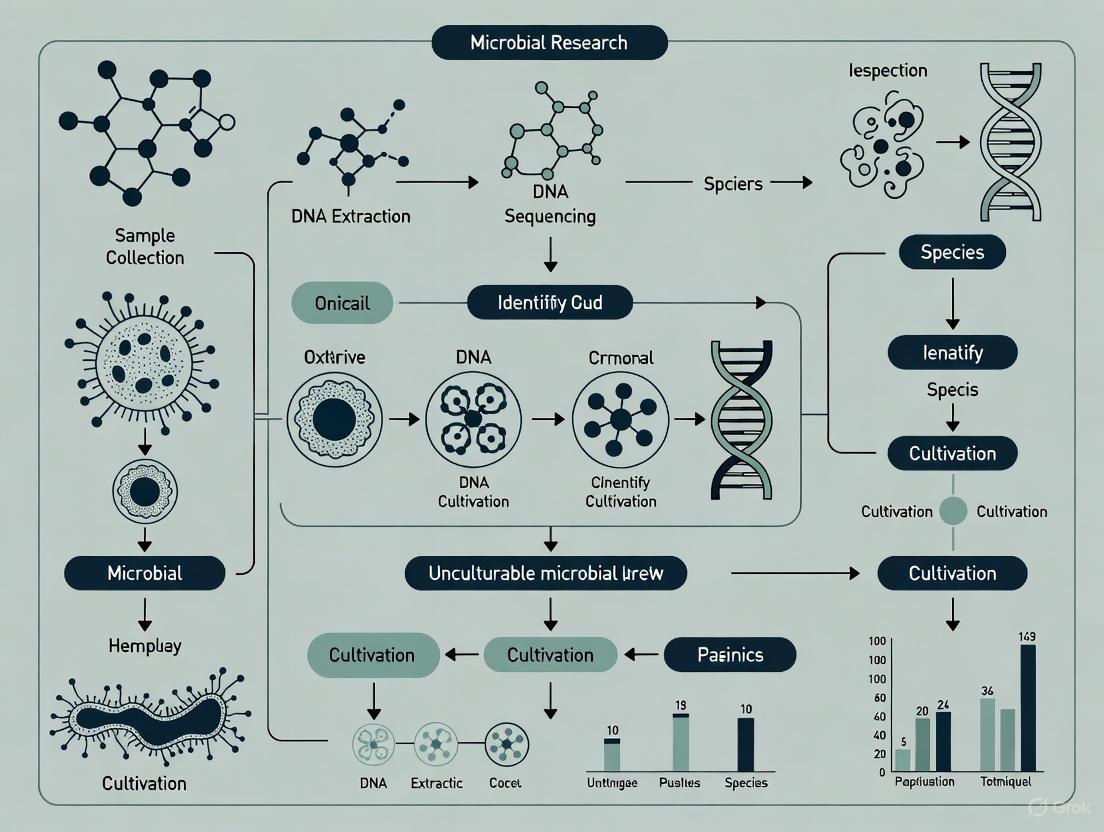
The workflow below visualizes the strategic approach to cultivating unculturable microorganisms, integrating both cultivation-independent and cultivation-dependent methods:
The table below summarizes the performance of various advanced cultivation techniques compared to traditional methods, demonstrating their efficacy in addressing the Great Plate Count Anomaly.
Table 1: Efficacy of Advanced Microbial Cultivation Techniques
| Cultivation Method | Typical Recovery Rate | Comparative Improvement Over Standard Plating | Key Isolates / Outcomes | Reference |
|---|---|---|---|---|
| Standard Plating | 0.01% - 1% of total cells | (Baseline) | Commonly culturable genera | [1] [5] |
| Diffusion Chambers (in situ) | Up to 40% of total cells | 14- to 1,400-fold | Previously uncultured marine bacteria | [1] |
| High-Throughput Extinction Culturing | Up to 14% of total cells | 14- to 1,400-fold | SAR11, OM43, SAR92 clades | [5] |
| Novel Diffusion Bioreactor (Soil) | 35 previously uncultured strains | Significantly higher than conventional method | Uncultured soil bacteria from four phyla | [4] |
| Co-culture with Helper Strains | Increased diversity | 51 novel species with Rpf factor | Increased taxonomic diversity from soil | [6] |
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagents and Materials for Cultivation of Unculturable Microorganisms
| Item | Function / Application | Example / Specification |
|---|---|---|
| Low-Nutrient Media | To cultivate oligotrophic bacteria that are inhibited by standard, rich media. | Dilute Peptone (0.01%), R2A (full or 50% strength), unamended filter-sterilized environmental water. |
| Environmental Extracts | To introduce native nutrients, trace elements, and potential signaling molecules into the growth medium. | Soil Extract, Water Extract (filter-sterilized through a 0.2 µm membrane). |
| Alternative Gelling Agents | To provide a solid surface without potential inhibitory compounds found in agar. | Gellan Gum. |
| Semi-Permeable Membranes | The core component of diffusion-based techniques, allowing chemical exchange while containing cells. | Polycarbonate Membranes (0.4 µm or 0.2 µm pore size). |
| Microtiter Plates | For high-throughput extinction culturing and co-culture experiments. | 48-well or 96-well non-tissue-culture-treated plates. |
| Resuscitation-Promoting Factor (Rpf) | A bacterial cytokine that can stimulate the resuscitation of dormant cells and promote growth. | Purified Rpf or culture supernatant from Micrococcus luteus [6]. |
| Stable Isotopes (for SIP) | To label and subsequently isolate DNA from active microorganisms within a community. | ¹³C-labeled or ¹âµN-labeled substrates for Stable Isotope Probing (SIP) [7]. |
| MMBC | MMBC, MF:C20H13NO7, MW:379.3 g/mol | Chemical Reagent |
| 5-HT2A receptor agonist-6 | 5-HT2A receptor agonist-6, CAS:1028307-48-3, MF:C18H19N3O3, MW:325.4 g/mol | Chemical Reagent |
The vast majority of the microbial world remains a scientific frontier, with an estimated 99% of microorganisms eluding standard laboratory cultivation techniques [9]. This "microbial dark matter" represents an enormous reservoir of unexplored biological diversity and genetic potential. Current genomic analyses reveal that of the 194 bacterial and archaeal phyla identified through the Genome Taxonomy Database, a staggering 85 phyla lack any cultured representatives [10]. This cultivation gap severely limits our understanding of microbial evolution, physiology, and ecology, while simultaneously restricting access to novel compounds with potential biotechnological and therapeutic applications.
The following technical support content is designed to assist researchers in overcoming the fundamental challenges in cultivating previously uncultured microorganisms. By integrating modern genomic insights with refined cultivation methodologies, this resource provides practical frameworks for bringing elusive microbial taxa into culture.
FAQs: Understanding Microbial Cultivation Challenges
1. Why are so many microorganisms considered "unculturable" using standard methods?
Most environmental microbes possess physiological traits and growth requirements that are not met by conventional cultivation approaches [11]. Key factors include:
- Unknown growth requirements: Many uncultured organisms require specific nutrients, signaling molecules, or growth factors that are not present in standard media [12]
- Slow growth rates: Oligotrophic (low-nutrient adapted) microorganisms grow slowly and are easily outcompeted by fast-growing copiotrophs in traditional rich media [10]
- Dependency on other organisms: Many microbes require metabolic byproducts from other community members through cross-feeding relationships [13]
- Low abundance: Some species exist at very low cell densities in their environments, making them difficult to enrich and isolate [13]
- Environmental sensitivity: Some organisms may be sensitive to oxygen, light, or other stressors encountered during standard cultivation attempts [12]
2. How can genomic data help us cultivate previously uncultured microorganisms?
Metagenomic and single-cell genomic information provides critical insights for designing targeted cultivation strategies [13] [14]:
- Metabolic pathway reconstruction: Genomic data can reveal an organism's nutritional requirements, energy metabolism, and potential auxotrophies [13]
- Habitat simulation: Genomic information about habitat conditions (temperature, pH, salinity) allows researchers to better mimic natural environments [11]
- Growth factor identification: Analysis can identify requirements for specific vitamins, cofactors, or signaling molecules [12]
- Community interaction mapping: Genomes can reveal dependencies on other microorganisms, guiding co-culture approaches [13]
3. What is the difference between oligotrophs and copiotrophs, and why does it matter for cultivation?
Table: Characteristics of Oligotrophic vs. Copiotrophic Microorganisms
| Characteristic | Oligotrophs | Copiotrophs |
|---|---|---|
| Nutrient Adaptation | Low nutrient concentrations | High nutrient concentrations |
| Growth Rate | Slow | Fast |
| Representation in Culture Collections | Underrepresented | Overrepresented |
| Environmental Abundance | Often high | Often low |
| Cultivation Media | Low nutrient media required | Grow on standard rich media |
Understanding this distinction is critical because most cultivation efforts historically have favored copiotrophs, creating a significant bias in microbial culture collections [10]. The "great plate count anomaly" refers to the observation that typically only 1% of environmental microbes form colonies on standard agar plates, primarily because most are oligotrophs inhibited by rich media [10].
4. What are the most promising new technologies for cultivating uncultured microbes?
Several innovative approaches have shown significant success:
- High-throughput dilution-to-extinction: Minimizes competition by separating individual cells in low-nutrient media [10]
- Reverse genomics: Using genetic sequences to design targeted capture and cultivation methods [15]
- Microfluidic devices: Enabling single-cell isolation and culture under controlled conditions [14]
- In situ cultivation: Allowing microorganisms to grow in their natural environment before laboratory transfer [14]
- Co-culture systems: Cultivating multiple species together to satisfy cross-feeding requirements [13]
Troubleshooting Guides
Problem: Inability to Isate Slow-Growing or Low-Abundance Species
Symptoms:
- Fast-growing species consistently overgrow plates
- No growth observed despite confirmed viable cells in environmental samples
- Small colonies that stop growing after reaching microscopic size
Solutions:
- Apply high-throughput dilution-to-extinction cultivation [10]
- Use 96-well plates with low-nutrient media
- Dilute inoculum to approximately 1 cell per well
- Incubate for extended periods (6-8 weeks or longer)
- Use multiple defined media formulations mimicking natural conditions
Reduce nutrient concentrations in media [12]
- Use carbon concentrations at micromolar levels
- Avoid rich organic substrates like yeast extract or peptone
- Consider artificial media that mimic natural water or soil solutions
Extend incubation times significantly beyond standard protocols [12]
- Some oligotrophic microorganisms require 30+ days to form visible colonies
- Protect plates from desiccation during extended incubation
- Monitor growth using molecular methods (PCR) rather than visual inspection
Problem: Contamination or Overgrowth by Fast-Growing Species
Symptoms:
- Same rapidly-growing species appear regardless of media used
- Unable to obtain pure cultures of target organisms
- Molecular analysis reveals diverse species in initial inoculation that disappear after transfer
Solutions:
- Use selective physical treatments
- Pre-filtration through different pore-size filters to select for specific size fractions
- Mild heat treatment to select for spore-formers or heat-resistant species
- Centrifugation gradients to separate cells by density
Apply chemical selection methods
- Incorporate antibiotics targeted against common contaminants
- Use specific carbon sources that only support growth of desired taxa
- Incorporate humic acids or other natural compounds that stimulate target organisms [12]
Modify atmospheric conditions [12]
- Reduce oxygen concentrations (1-2% Oâ‚‚) for microaerophiles
- Increase COâ‚‚ concentrations (5%) for capnophiles
- Create anoxic conditions for strict anaerobes
Problem: Failure to Replicate Natural Environmental Conditions
Symptoms:
- Cells grow initially but cannot be subcultured
- Organisms grow in environmental samples but not in laboratory media
- Growth occurs only in mixed culture but not in isolation
Solutions:
- Incorporate chemical signals and growth factors [12]
- Add acyl homoserine lactones or other quorum-signaling molecules
- Include catalase or pyruvate to detoxify hydrogen peroxide [12]
- Add humic acids or analogs like anthraquinone disulfonate
Simulate natural substrate concentrations
- Use carbon sources at environmentally relevant concentrations (μg/L to mg/L range)
- Replicate the stoichiometry of natural waters or soils
- Include organic nitrogen and phosphorus sources at appropriate ratios
Implement co-culture approaches [13]
- Culture with helper strains that provide essential nutrients
- Use feeder layers or conditioned media from other cultures
- Create synthetic communities of 2-3 species rather than pursuing pure culture
Experimental Protocols
Protocol 1: High-Throughput Dilution-to-Extinction Cultivation for Freshwater Oligotrophs
Based on successful isolation of 627 axenic strains from freshwater ecosystems [10]
Research Reagent Solutions:
Table: Essential Reagents for Dilution-to-Extinction Cultivation
| Reagent | Function | Application Notes |
|---|---|---|
| Defined oligotrophic media (med2/med3) | Mimics natural freshwater nutrient conditions | Contains carbohydrates, organic acids, vitamins in μM concentrations |
| MM-med medium | Selects for methylotrophic bacteria | Contains methanol, methylamine as sole carbon sources |
| Catalase | Detoxifies hydrogen peroxide | Critical for preventing oxidative stress during isolation |
| SMRTbell prep kit 3.0 | Library preparation for genome sequencing | Enables verification of isolates via whole genome sequencing [16] |
| Barcoded adapter plates | Multiplexing samples for sequencing | Allows efficient genome sequencing of multiple isolates [16] |
Methodology:
- Sample collection and processing:
- Collect water samples from appropriate depth (epilimnion vs. hypolimnion)
- Process within 24 hours of collection with storage at 4°C
- Pre-filter through 3μm filters to remove larger organisms and particles
Inoculation and incubation:
- Prepare serial dilutions in multiple defined media types
- Dispense into 96-deep-well plates at approximately 1 cell per well
- Incubate at in situ temperatures (16°C for temperate lakes) for 6-8 weeks
- Monitor growth visually and via molecular screening
Molecular screening and isolation:
- Screen using PCR-based methods like plate wash PCR (PWPCR) [12]
- Identify wells containing target taxa via 16S rRNA gene sequencing
- Transfer positive cultures to fresh media for purification
- Confirm axenic status by microscopy and repeated molecular analysis
Protocol 2: Metagenome-Guided Targeted Isolation
Methodology:
- Metagenomic analysis:
- Sequence environmental metagenomes from target habitat
- Reconstruct metagenome-assembled genomes (MAGs) for uncultured taxa
- Analyze metabolic pathways to predict growth requirements [13]
Media design based on genomic predictions:
- Include carbon sources identified in metabolic reconstructions
- Add required vitamins or cofactors based on identified auxotrophies
- Adjust oxygen conditions based on detected respiratory pathways
- Incorporate potential electron donors/acceptors
Targeted isolation:
- Use fluorescence-activated cell sorting with designed probes
- Apply antibody-based capture for specific cell types
- Use diffusion chambers or in situ devices to simulate natural conditions
Quantitative Context: The Scale of Microbial Diversity
Table: Quantifying the Uncultured Microbial Majority
| Diversity Metric | Value | Source/Context |
|---|---|---|
| Estimated global prokaryotic diversity | 2.2-4.3 million operational taxonomic units | Based on 16S rRNA gene surveys [13] |
| Bacterial phyla with no cultured representatives | 85 phyla | From 194 total phyla in GTDB [10] |
| Cultured vs. validly described species | 24,745 described of 113,104 species clusters | GTDB Release R220 [10] |
| Uncultured genera and phyla in environmental samples | 81% and 25% of microbial cells | Across Earth's microbiomes [13] |
| Success rate with high-throughput dilution-to-extinction | 10 axenic strains per sample (12.6% viability) | Freshwater cultivation study [10] |
The systematic cultivation of previously uncultured microorganisms requires moving beyond traditional methods to embrace integrated approaches that combine genomic insights with refined cultivation techniques. By implementing the troubleshooting guides, experimental protocols, and methodological frameworks presented in this technical support resource, researchers can significantly advance their ability to bring elusive microbial taxa into culture. Each new cultivated isolate not only expands our understanding of microbial diversity but also provides opportunities for discovering novel metabolic capabilities, ecological interactions, and potentially valuable compounds for biotechnology and medicine. The cultivation tools now available, particularly when guided by genomic data, are progressively dismantling the concept of "uncultivability" that has long limited microbial discovery.
The overwhelming majority of microorganisms in nature have evaded traditional laboratory cultivation, creating a significant bottleneck in the discovery of novel antibiotics and natural products. It is estimated that less than 1% of bacterial diversity can be cultured using standard techniques, leaving over 99% of microorganisms, often referred to as "microbial dark matter," unexplored [17] [4]. This cultivation gap coincides with a critical "discovery void" in antibiotic development; the most recent novel class of antibiotics to reach the market was discovered in 1987 [18] [19] [20]. Overcoming the technical challenges of growing uncultured species is therefore not merely an academic exercise, but a crucial endeavor for accessing new chemical scaffolds to combat the global antimicrobial resistance (AMR) crisis [1] [21]. This guide provides targeted troubleshooting and methodologies to help researchers access this untapped reservoir of biodiversity.
FAQs: Core Concepts and Common Challenges
Q1: What does it mean for a microorganism to be "unculturable"?
The term "unculturable" is a misnomer; it indicates that current standard laboratory culturing techniques are unable to support the growth of a given bacterium. These organisms are clearly metabolically active and growing in their natural environment, but we lack critical information about their specific biological and nutritional requirements to replicate those conditions in the lab [1]. The inability to cultivate them signifies a gap in our knowledge, not an inherent property of the microbe itself [4].
Q2: Why is accessing uncultured bacteria so critical for new antibiotic discovery?
Historically, natural products from bacteria have been the source of over half of all commercially available pharmaceuticals [1]. The repeated isolation and screening of the same small fraction of culturable bacteria has led to a high rate of compound rediscovery and the collapse of the antibiotic discovery pipeline [1] [22]. It is estimated that to find a new class of antibiotics, more than 10 million isolates from known, culturable bacteria would need to be screened [1]. Accessing the uncultured majority dramatically expands the accessible genetic and metabolic diversity, offering the most promising path to discovering truly novel antibiotic classes [1] [17] [20].
Q3: What are the primary reasons these bacteria fail to grow in the lab?
The simple explanation is a failure to replicate essential aspects of their natural environment. The specific reasons are multifaceted and can include [1] [4]:
- Lack of essential nutrients or growth factors: The standard, nutrient-rich media may be inappropriate for oligotrophic (nutrient-poor environment-adapted) organisms.
- Absence of essential signaling molecules: Some bacteria require chemical signals from other microbes to initiate growth or division.
- Dependence on other organisms (helper strains): Some species rely on helper bacteria to provide specific metabolites, detoxify the environment, or remove oxidative stress.
- Incorrect physicochemical conditions: Factors such as pH, osmotic conditions, temperature, or oxygen tension may not be adequately replicated.
- Inhibition by standard media components: Some ingredients, like agar itself, can be inhibitory to certain microbes [4].
Troubleshooting Guide: Overcoming Cultivation Barriers
This section addresses specific experimental issues and offers solutions based on advanced cultivation strategies.
Table 1: Common Experimental Problems and Advanced Solutions
| Problem Symptom | Potential Root Cause | Recommended Solution | Key References |
|---|---|---|---|
| Very low recovery of novel isolates; overgrowth by fast-growing lab strains. | Standard media are too rich and selective for fast-growing, common bacteria. | Use low-nutrient media (e.g., 1:10 or 1:100 dilution of R2A); incorporate soil extract (SE) to mimic natural conditions; use gellan gum as an alternative solidifying agent. | [4] |
| Suspected dependence on other microbes or signaling molecules. | The target organism requires growth factors provided by a helper community. | Implement co-culture techniques; use diffusion chambers or bioreactors that allow chemical exchange with a helper community or soil environment. | [1] [4] |
| No growth even after prolonged incubation. | Incubation time is insufficient for slow-growing oligotrophs. | Extend incubation time significantly to 4-8 weeks or longer; protect plates from desiccation during long-term incubation. | [4] |
| Inability to replicate the target organism's natural habitat. | Critical environmental parameters are not being mimicked in the lab. | Employ in situ cultivation using diffusion chambers incubated in the native environment, or use novel bioreactor designs that simulate the natural habitat. | [1] [4] |
| Known high biosynthetic potential from genetic data, but cultivation fails. | The biosynthetic gene clusters (BGCs) are silent under lab conditions. | Use metagenomics: construct large-insert libraries (e.g., BACs) from environmental DNA (eDNA) and express them in a heterologous host like E. coli or Streptomyces. | [17] [6] |
Experimental Protocols: Key Methodologies for Culturing Uncultured Bacteria
Protocol 1: Diffusion Bioreactor Cultivation
This protocol, adapted from an advanced cultivation study, uses a semi-permeable membrane to maintain cells in their natural chemical environment while allowing for the recovery of pure isolates [4].
1. Bioreactor Design and Setup:
- Construct a double-chambered system with an inner chamber placed inside an outer chamber.
- The inner chamber (e.g., a 2L plastic container) has walls drilled with ~160 holes (6mm diameter) and is covered on the outside with a polycarbonate membrane (0.4 µm pore size).
- Sterilize all components with 70% ethanol, UV light, and rinsing with particle-free water.
- Fill the gap between the inner and outer chamber with fresh, sieved soil from the sampling site.
- Add the soil inoculum (e.g., 3g) and liquid cultivation medium (e.g., 300mL of diluted R2A, R2A-SE, or J26-SE medium) to the inner chamber.
2. Cultivation and Isolation:
- Seal the bioreactor and incubate at room temperature with slow stirring for 4 weeks.
- After incubation, serially dilute the content from the inner chamber and spread onto agar plates of the same medium.
- Incubate the plates aerobically at 25°C for an additional 4 weeks.
- Periodically check for slow-growing microcolonies and purify them through repeated sub-culturing.
This method has been proven successful in cultivating previously uncultured strains by allowing continuous diffusion of essential nutrients and signaling molecules from the natural soil environment [4].
Protocol 2: Function-Based Metagenomic Workflow
This culture-independent protocol bypasses cultivation altogether to access biosynthetic gene clusters (BGCs) directly from environmental samples [17] [6].
1. Environmental DNA (eDNA) Extraction and Library Construction:
- Extract high-molecular-weight DNA directly from the environmental sample (e.g., soil, sediment). Indirect extraction methods that involve physical separation of cells first are preferred to obtain larger DNA fragments.
- Fragment the eDNA and clone it into a * Bacterial Artificial Chromosome (BAC) vector* to accommodate large gene clusters (up to 200 kb).
- Transform the constructed library into a heterologous host. While E. coli is common, consider alternative hosts like Streptomyces lividans, Pseudomonas putida, or Ralstonia metallidurans for better expression of genes from phylogenetically distant bacteria [17].
2. Screening for Natural Product Biosynthesis:
- Function-based screening: Screen library clones for desired antimicrobial activity against indicator strains or for specific phenotypes (e.g., pigment production).
- Sequence-based screening: Alternatively, screen the library using next-generation sequencing or PCR for conserved biosynthetic genes like Polyketide Synthases (PKS) and Non-Ribosomal Peptide Synthetases (NRPS) [17] [6].
The following diagram illustrates the logical workflow for this metagenomic approach, showing the two primary screening paths.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Reagent Solutions for Cultivation and Metagenomics
| Reagent / Material | Function / Application | Key Considerations |
|---|---|---|
| Soil Extract (SE) | A component of culture media to simulate the chemical environment of soil, providing unknown essential nutrients and trace elements. | Prepare from the same habitat as the target inoculum to increase relevance [4]. |
| Gellan Gum | An alternative solidifying agent to agar. | Can prevent the inhibition of growth observed in some fastidious microbes when agar is used [4]. |
| R2A Medium (Diluted) | A low-nutrient medium ideal for isolating slow-growing oligotrophic bacteria from water and soil. | Diluting to 10% or 50% of standard strength can further improve recovery of novel isolates [4]. |
| Diffusion Chamber / Bioreactor | A device that allows chemical exchange between the native environment and the cultivation chamber, mimicking natural habitat. | Can be custom-built or use commercial Transwell plates. Critical for supplying unknown growth factors [1] [4]. |
| Bacterial Artificial Chromosome (BAC) Vector | A cloning vector capable of maintaining very large inserts of foreign DNA (100-200 kb), suitable for entire biosynthetic gene clusters. | Essential for function-based metagenomics to capture large operons for natural product synthesis [17]. |
| Heterologous Hosts (e.g., S. lividans) | Expression systems for eDNA libraries. Alternative hosts can improve expression of BGCs compared to the standard E. coli host. | Matching the phylogenetic background of the eDNA and the host can dramatically increase success rates [17]. |
| Resuscitation-Promoting Factor (Rpf) | A bacterial cytokine that stimulates the growth of dormant and recalcitrant bacteria. | Adding Rpf or culture supernatant containing it (e.g., from Micrococcus luteus) can increase microbial diversity in cultures [6]. |
| DL-Tyrosine-d7 | L-4-Hydroxyphenyl-D4-alanine-2,3,3-D3 | L-4-Hydroxyphenyl-D4-alanine-2,3,3-D3 is a deuterated amino acid for metabolic and Parkinson's disease research. For Research Use Only. Not for human use. |
| 14-Norpseurotin | 14-Norpseurotin, CAS:1031727-34-0, MF:C21H23NO8, MW:417.4 g/mol | Chemical Reagent |
Troubleshooting Guide for Microbial Cultivation
Cultivating environmentally relevant microorganisms, particularly oligotrophs and interdependent species, presents unique challenges. This guide addresses common failure points and provides targeted solutions to bring the "uncultivated microbial majority" into culture [10] [23].
Problem: No Growth or Extremely Slow Growth of Inoculant
Potential Cause & Solution:
Potential Cause Recommended Action Key References / Rationale Inappropriate nutrient concentration (Media is too rich for oligotrophs) [10]. Use low-nutrient media or dilution-to-extinction techniques with defined media that mimic natural substrate concentrations [10] [24]. Rationale: Oligotrophs are inhibited by high nutrient levels. Success was shown using media with 1.1-1.3 mg DOC/L [10]. Insufficient incubation time [24]. Extend incubation periods significantly—weeks or months—instead of days [24]. Rationale: Slow-growing oligotrophs require extended time. Isolates from Antarctic soil appeared after 4-15 weeks [24]. Missing essential growth factors or vitamins [10] [23]. Supplement media with specific growth factors like zincmethylphyrins, short-chain fatty acids, or vitamins based on genomic/metagenomic predictions [23]. Rationale: Many aquatic prokaryotes have reduced genomes with multiple auxotrophies [10].
Problem: Contamination by Fast-Growing Copiotrophs
Potential Cause & Solution:
Potential Cause Recommended Action Key References / Rationale Fast-growing copiotrophs outcompete target slow-growers [10]. Apply high-throughput dilution-to-extinction cultivation, inoculating at approximately one cell per well to isolate slow-growers from competitors [10]. Rationale: This method physically separates target cells, preventing overgrowth. 627 axenic strains were isolated this way [10]. Inadequate sterilization of equipment or media. Implement rigorous sterilization protocols and work in a sterile environment, ideally with a laminar flow hood [24]. Rationale: Critical for excluding contaminants during long incubations.
Problem: Growth Cessation Upon Subculturing
Potential Cause & Solution:
Potential Cause Recommended Action Key References / Rationale Dependence on other microbes for essential nutrients or detoxification [10] [23]. Employ co-cultivation strategies or use diffusion chambers/iCHIP devices that allow chemical exchange with the native environment [23]. Rationale: Dependencies on co-occurring microbes are common. Co-culture helped isolate Candidatus Prometheoarchaeum syntrophicum [23]. Uncharacterized dependency on signaling molecules. Experiment with conditioned media from environmental samples or known microbial partners. Rationale: Intraspecific and interspecies interactions (e.g., quorum sensing) profoundly affect growth [23].
Frequently Asked Questions (FAQs)
Q1: What are the most effective media strategies for isolating oligotrophic microbes? The most successful strategies involve using defined, artificial media with low carbon content (e.g., 1.1-1.3 mg DOC/L) that closely mimics the chemical composition of the natural environment from which the sample was taken. This avoids the inhibition of oligotrophs that occurs on standard nutrient-rich media [10]. Dilution-to-extinction in such media is a highly effective high-throughput method [10].
Q2: How long should I wait for growth to appear before discarding a culture? For many uncultured oligotrophs, incubation periods need to be significantly longer than standard protocols. Viable cultures can appear after 4 weeks, and some may require up to 15 weeks of incubation. Patience and measures to prevent media desiccation are crucial [24].
Q3: My isolated strain grows poorly in liquid media but better on solid surfaces. Why? Some slow-growing environmental bacteria, particularly those with a free-living lifestyle, show a preference for solid surfaces or may form microcolonies that are difficult to disperse in liquid. Continued cultivation on solid media, such as gellan gum-based plates, is often a viable solution [10] [24].
Q4: How can genomic data inform my cultivation strategies? Genome-centric metagenomics and single-cell genomics can reveal metabolic deficiencies (auxotrophies) and potential metabolic pathways of your target organism. This information allows for the rational design of specific cultivation media by supplementing missing vitamins, cofactors, or specific carbon sources that the microbe is predicted to require [23].
Experimental Protocols for Key Methodologies
Protocol 1: High-Throughput Dilution-to-Extinction Cultivation
This protocol is adapted from methods that successfully isolated 627 axenic strains of abundant freshwater bacteria [10].
- Objective: To isolate slow-growing oligotrophic microbes by physically separating them from faster-growing competitors and providing a low-nutrient environment.
- Materials:
- Sterile, defined oligotrophic media (e.g., med2 or med3 from reference [10])
- 96-deep-well plates
- Environmental sample (water, soil suspension)
- Sterile dilution blanks
- Procedure:
- Sample Preparation: Serially dilute the environmental sample in a sterile, dilute saline solution (e.g., 0.9% NaCl) [24].
- Inoculation: Inoculate 96-deep-well plates with a dilution calculated to contain approximately one microbial cell per well [10].
- Incubation: Incubate plates at a temperature relevant to the sample's origin (e.g., 16°C for freshwater lakes) for 6-8 weeks or longer [10].
- Screening: Monitor wells for turbidity. Screen positive wells for purity via 16S rRNA gene sequencing.
- Purification: Transfer cultures from positive wells to fresh defined media to obtain axenic strains.
Protocol 2: Cultivation Using Long Incubation and Low Nutrients
This protocol is effective for isolating rare and slow-growing microorganisms from soil and other solid samples [24].
- Objective: To isolate recalcitrant microorganisms by simulating low-nutrient conditions and allowing for extremely slow growth rates.
- Materials:
- Low-nutrient media (e.g., 1/100 diluted Nutrient Broth, ~0.08 g/L) [24]
- Gellan gum (as a solidifying agent)
- MgSO₄·7H₂O
- Procedure:
- Media Preparation: Prepare 1/100 diluted Nutrient Broth, solidified with 0.7% gellan gum. Add 0.1% MgSO₄·7H₂O to promote solidification [24].
- Plating: Spread plate serial dilutions of the soil suspension onto the prepared media.
- Incubation: Incubate plates at a low temperature (e.g., 12°C) in sealed polyethylene bags to prevent desiccation for up to 15 weeks [24].
- Selection: Weekly, use a stereo microscope to identify and mark new, morphologically distinct colonies that appear after 4 weeks of incubation.
- Isolation: Pick and streak these late-appearing colonies onto fresh plates of the same low-nutrient media to obtain pure isolates.
Workflow Visualization
The following diagram illustrates a logical troubleshooting workflow for addressing microbial cultivation failures.
The Scientist's Toolkit: Research Reagent Solutions
Essential materials and reagents for advanced cultivation of uncultured microbes.
| Item | Function & Application |
|---|---|
| Defined Oligotrophic Media (e.g., med2/med3 [10]) | Provides a reproducible, low-nutrient environment that mimics natural conditions to avoid inhibiting oligotrophs. |
| Gellan Gum | A superior solidifying agent for plates, often preferred over agar for the growth of certain environmental microbes [24]. |
| Diffusion Chambers / iCHIP [23] | Devices that allow microbes to be cultivated in situ by permitting the diffusion of environmental nutrients and signals. |
| Specific Growth Factors (e.g., vitamins, short-chain fatty acids, coproporphyrins [23]) | Supplements to address auxotrophies predicted by genomic analysis or suspected in fastidious microbes. |
| Selective Inhibitors (e.g., diuron [23]) | Used to suppress the growth of oxygenic phototrophs or other competing microorganisms in enrichment cultures. |
| 3-Hydroxy-3-methylglutaryldithio-CoA | 3-Hydroxy-3-methylglutaryldithio-CoA, CAS:134785-93-6, MF:C27H44N7O20P3S2, MW:943.7 g/mol |
| 3-Hydroxystearic acid | 3-Hydroxystearic acid, CAS:45261-96-9, MF:C18H36O3, MW:300.5 g/mol |
Cultivation Breakthroughs: From Simulated Environments to High-Throughput Technologies
Troubleshooting Guide: Common Experimental Issues and Solutions
This guide addresses specific, frequently encountered problems when performing dilution-to-extinction cultivation, helping to ensure the successful isolation of previously unculturable species.
Table: Troubleshooting Common Issues in Dilution-to-Extinction Cultivation
| Problem | Probable Cause | Recommended Solution | Prevention Tip |
|---|---|---|---|
| No bacterial growth in wells after incubation [25] | Over-dilution of the bacterial suspension. | Reduce the dilution factor or increase the starting inoculum concentration [25]. | Perform a pilot test to determine the optimal dilution for your sample. |
| Excessive growth in all wells [25] | Bacterial concentration in the dilution series is too high. | Prepare a more diluted suspension before plating to optimize for microbial diversity [25]. | Use cell counting (e.g., flow cytometry) to standardize the initial inoculum [10]. |
| Cross-contamination between wells [25] | Splashing during pipetting. | Use slow and controlled pipetting; centrifuge the plate before pipetting to minimize splashing [25]. | Work in a laminar flow hood and use filter tips. |
| Drying of liquid medium in outer wells [25] | Incomplete sealing of microplates. | Ensure plates are tightly sealed with Parafilm, paying special attention to the edges [25]. | Use plates with low-evaporation lids and incubate in a humidified chamber. |
| No visible PCR product [25] | PCR inhibitors from the sample or degraded DNA. | Dilute the DNA template 1:10 or purify it using a cleanup kit [25]. Avoid overheating during alkaline lysis [25]. | Include a positive control in the PCR reaction. |
| Low recovery rate after freezing [25] | Sensitivity to freeze-thaw process; insufficient cryoprotectant mixing. | Increase bacterial concentration in glycerol stocks; avoid repeated freeze-thaw cycles [25]. | Ensure glycerol and bacterial culture are thoroughly mixed before storage at -80°C [25]. |
Frequently Asked Questions (FAQs)
1. What is the core principle behind dilution-to-extinction cultivation, and why is it effective for "unculturable" species?
Dilution-to-extinction involves progressively diluting a microbial sample in a growth medium until, theoretically, only a single cell remains in some of the wells [26]. This method is effective for isolating rare and slow-growing bacteria for two main reasons. First, it physically separates cells, preventing fast-growing "weeds" from outcompeting slow-growing or rare taxa [25] [27]. Second, it allows researchers to use very low-nutrient defined media that mimic a microbe's natural oligotrophic environment, which is often essential for the growth of many environmentally relevant microbes that are inhibited by standard rich media [10] [28].
2. How do I choose between a defined medium and a natural medium like autoclaved lake water?
Both approaches have merits. Defined artificial media are preferred for reproducibility and for systematically determining the specific nutritional requirements of an isolate [10]. They allow for precise control over every component. Natural media (e.g., filter-sterilized lake or seawater) can be highly effective as they inherently contain the natural mix of nutrients and trace elements from the environment [29]. However, their composition can vary seasonally, and sterilization can degrade heat-sensitive compounds [10]. A modern strategy is to create defined media that chemically mimic natural conditions, often containing µM concentrations of carbon sources, vitamins, and other organic compounds [10].
3. Our team successfully isolated a novel bacterium using this method, but it grows very slowly. How can we improve its growth?
Slow growth is a common characteristic of many oligotrophic isolates [10]. To improve growth, consider:
- Co-cultivation: Some bacteria depend on other microbes for essential nutrients or detoxification of metabolites [1] [10]. Try growing your isolate with a helper strain from the same environment.
- Supplementation: Add small amounts of catalase to degrade toxic reactive oxygen species, which has been key to cultivating groups like the freshwater acI Actinobacteria [30]. Other supplements like resuscitation-promoting factor (Rpf) can also stimulate growth [6].
- Patience: Ensure your incubation times are sufficiently long (weeks to months) and avoid frequent sub-culturing, which can stress slow-growing organisms [1].
4. Our sequencing results show low-quality reads or unequal depth across samples. What went wrong?
This is often a problem with the PCR step prior to sequencing. Low-quality reads can result from poor primer specificity or degraded PCR products [25]. To fix this, use a high-fidelity polymerase and verify your primer design. Unequal sequencing depth is typically caused by uneven concentrations of PCR products before pooling [25]. Always double-check and normalize PCR product concentrations to ensure an even representation of all samples [25].
5. What are the major limitations of high-throughput dilution-to-extinction cultivation?
While powerful, this method has inherent biases:
- Liquid Medium Bias: It preferentially isolates microbes that grow well in liquid, potentially missing those that require solid surfaces for biofilm formation [25].
- Aerobic Bias: The protocol is typically designed for aerobic conditions and will miss obligate anaerobes without modification [25].
- Loss of Interdependent Microbes: The method isolates individual cells, disrupting syntrophic or symbiotic relationships essential for some species [25] [1].
- Medium Selectivity: The choice of medium (e.g., Tryptic Soy Broth) will favor certain groups while excluding oligotrophs or extremophiles that require specialized nutrients [25] [28].
Experimental Workflow and Methodology
The following diagram illustrates the key stages of a high-throughput dilution-to-extinction cultivation experiment, from sample preparation to the generation of a culture collection.
Core Protocol Steps:
- Sample Collection & Preparation: Fresh environmental samples (e.g., plant roots, lake water) are collected and processed to create a homogeneous cell suspension in a buffer like sterile phosphate-buffered saline (PBS) [25] [27].
- Serial Dilution & Dispensing: The cell suspension is subjected to a series of dilutions in a defined, low-nutrient culture medium. The diluted suspension is then dispensed into the wells of 96-well plates, with a theoretical dilution aiming for one cell per well [25] [10].
- Incubation: Plates are sealed to prevent evaporation and incubated for extended periods (from several weeks to months) at a temperature reflective of the natural environment [27] [10]. This allows slow-growing oligotrophs to proliferate.
- Growth Screening & Purification: Wells are visually inspected for turbidity, indicating microbial growth [27]. Positive wells are transferred to fresh medium to ensure purity and robust growth. Purity is confirmed via microscopy and sequencing.
- Identification & Preservation: Isolates are identified using 16S rRNA gene sequencing, and pure cultures are preserved in glycerol stocks at -80°C for long-term storage and future studies [25] [10].
The Scientist's Toolkit: Key Research Reagents and Materials
Table: Essential Reagents for Dilution-to-Extinction Cultivation with Defined Media
| Reagent/Material | Function/Application | Example from Literature |
|---|---|---|
| Defined Oligotrophic Media | Simulates natural nutrient conditions to grow bacteria adapted to low nutrient availability (oligotrophs) [10]. | Artificial freshwater media (e.g., med2, med3) with µM concentrations of carbon sources, vitamins, and organic compounds [10]. |
| Catalase | Degrades toxic hydrogen peroxide, a common stressor in vitro. Crucial for isolating sensitive taxa. | Supplementation was key to cultivating previously uncultured acI lineage Actinobacteria from a lake [30]. |
| Resuscitation-Promoting Factor (Rpf) | A bacterial growth factor that stimulates the resuscitation of dormant cells from a viable but non-culturable state. | Micrococcus luteus culture supernatant containing Rpf increased the diversity of cultured bacteria from soil [6]. |
| Complex Natural Carbon Sources | Provides a diverse mixture of organic compounds to support the growth of bacteria with unknown or complex nutritional needs. | Sediment dissolved organic matter (DOM) and bacterial cell lysate were far more effective than simple sugars in cultivating diverse subsurface microbes [28]. |
| High-Throughput Screening Tools | Enables efficient processing of thousands of cultures. | Use of 96-well deep-well plates, automated liquid handling, and flow cytometry for growth screening [25] [29]. |
| Magnetic Beads for DNA Clean-up | Purifies PCR products by removing contaminants and enzymes, essential for high-quality sequencing library preparation. | Mag-Bind TotalPure NGS magnetic beads used to clean up PCR products before sequencing [25]. |
| 2'-Deoxy-NAD+ | 2'-Deoxy-NAD+, CAS:151411-04-0, MF:C21H28N7O14P2+, MW:664.4 g/mol | Chemical Reagent |
| Quifenadine hydrochloride | Quifenadine Hydrochloride|CAS 10447-38-8|H1 Blocker |
The pursuit of cultivating the "unculturable" represents one of the most significant challenges in modern microbiology. The vast majority of microbial species on Earth resist growth under standard laboratory conditions, creating a substantial gap in our understanding of microbial diversity and its potential applications [1]. This technical support center is designed to equip researchers with practical methodologies for employing advanced in-situ cultivation techniques, specifically diffusion chambers and semi-permeable capsules. These approaches bridge the gap between the laboratory and the natural environment, allowing researchers to cultivate previously inaccessible microorganisms for drug discovery, ecological studies, and fundamental biological research [4] [31].
Key Experimental Protocols
Diffusion Bioreactor Assembly and Workflow
The diffusion bioreactor technique enables environmental nutrients and signalling molecules to reach encapsulated microorganisms, mimicking their natural habitat.
Detailed Methodology [4]:
- Chamber Construction: The system consists of two chambers. The inner chamber is a 2-L plastic container (approximately 140 mm wide × 150 mm tall). The outer chamber is a larger 4-L plastic container (approximately 240 mm wide × 120 mm tall).
- Membrane Preparation: Create approximately 160 holes (6 mm diameter) in the walls of the inner chamber. Affix a sterile polycarbonate membrane with a 0.4 µm pore size to the outer side of this chamber. This membrane allows for the diffusion of molecules while containing the cells.
- Sterilization: Sterilize all components with 70% ethanol, air-dry under UV light in a laminar flow hood for 24 hours, and perform a final rinse with particle-free molecular grade water.
- System Setup:
- Place the prepared inner chamber inside the outer chamber.
- Fill the gap between the two chambers with freshly collected and sieved environmental soil (e.g., forest soil) to recreate the natural chemical environment.
- Inside the inner chamber, add 3 grams of the soil sample to be cultured into 300 mL of a selected culture medium.
- Seal the inner chamber lid tightly with sealing tape.
- Incubation and Recovery: Incubate the entire bioreactor assembly at room temperature for up to 4 weeks with gentle stirring. After incubation, serially dilute the contents of the inner chamber, plate onto solid agar media, and incubate aerobically at 25°C for another 4 weeks to obtain pure isolates.
The following workflow diagram illustrates the key stages of this protocol:
Microencapsulation and In-Situ Incubation
This protocol uses agarose microbeads to encapsulate single cells or micro-colonies, protecting them and allowing for high-throughput analysis while exposed to their native environment.
Detailed Methodology [31]:
- Sample Preparation: Dislodge bacteria from the environmental sample (e.g., marine sediment). Wash the sediment with a diluted nutrient broth (e.g., 1:10 Marine Broth) to remove salts and debris. Concentrate the bacterial cells via centrifugation.
- Cell Encapsulation:
- Resuspend the concentrated cells in 1% (w/v) Low Gelling Temperature (LGT) agarose, prepared in a diluted culture medium and maintained at 35°C.
- Use a microfluidic chip to generate microbeads with a diameter of 80 ± 20 µm, containing the encapsulated bacteria.
- Dialysis Cassette In-Situ Incubation:
- Load the agarose microbeads (or a non-encapsulated cell resuspension for comparison) into a modified dialysis cassette (e.g., a Slyde-A-Lyzer cassette).
- Seal the cassette and incubate it in the original natural environment (e.g., submerged in the source marine water) for one week. This allows for continuous diffusion of environmental compounds.
- Laboratory Cultivation: After the in-situ incubation, retrieve the cassette. Plate the contents (either the microbeads or the liquid suspension) onto solid culture media and incubate under appropriate laboratory conditions to recover isolates.
Troubleshooting Guides
Low Recovery of Novel Isolates
| Symptom | Possible Cause | Solution |
|---|---|---|
| Low diversity of recovered colonies; predominantly fast-growing, common species. | Nutrient-rich media favor fast-growing bacteria and inhibit oligotrophic (nutrient-preferring) species [1]. | Use low-nutrient media such as 50% diluted R2A, or media supplemented with soil extract (SE) or habitat-specific extracts [4]. |
| Incubation time is too short for slow-growing species to form visible colonies [4]. | Extend incubation periods significantly, up to several weeks or months [4]. | |
| Missing essential growth factors or signaling molecules provided by other microbes in the natural community [1]. | Employ co-culture techniques with "helper" strains from the same environment, or use diffusion systems that allow exchange of metabolites [1]. |
Contamination and System Integrity
| Symptom | Possible Cause | Solution |
|---|---|---|
| Growth of contaminants (e.g., mold, common lab bacteria) in the inner chamber or capsules. | Compromised membrane or seal, allowing invasive cells to enter [32]. | Prior to use, perform an overpressure test on assembled chambers to check for leaks. Replace O-rings and seals regularly [32]. |
| Contaminated inoculum or laboratory media [32]. | Check the sterility of the inoculum by plating a sample on a rich medium. Ensure all media and reagents are properly sterilized [32]. | |
| Contamination introduced during the assembly process. | Pre-assemble as many components as possible before sterilization. Minimize connections made after autoclaving [32]. |
Frequently Asked Questions (FAQs)
Q1: What is the main advantage of using an in-situ diffusion chamber over traditional lab plates? A1: Diffusion chambers and capsules allow microorganisms to be exposed to the natural chemical and physical gradients of their original habitat, including nutrients, signaling molecules, and metabolic byproducts from neighboring organisms. This simulated natural environment provides essential growth conditions that are impossible to replicate with synthetic media in a petri dish, enabling the cultivation of previously "unculturable" species [1] [4].
Q2: How does the pore size of the membrane affect the experiment? A2: The membrane must have a pore size small enough to physically contain the bacterial cells (typically 0.4 µm or smaller) but large enough to allow the free diffusion of nutrients, dissolved gases, and other critical small molecules from the external environment into the chamber [4].
Q3: We are not recovering any growth after retrieval. What could be wrong? A3: First, verify that your sterilization process (e.g., autoclaving) reached the correct temperature and time, and that steam could penetrate all parts of the equipment [32]. Second, ensure the membrane is not clogged with soil particles, which would prevent diffusion. Third, consider extending the in-situ incubation period, as some slow-growing species may require more time to initiate replication [4].
Q4: Can these techniques be used for anaerobic microorganisms? A4: Yes, the principle is adaptable. The system must be assembled and incubated in an anaerobic environment to ensure that oxygen does not diffuse through the membrane and into the chamber. This typically requires the use of an anaerobic chamber for setup and placement in an anoxic environment for incubation [33].
Data Presentation: Optimizing Recovery
The following table summarizes quantitative findings from a key study that utilized a diffusion bioreactor for cultivating soil bacteria, highlighting the impact of critical experimental parameters [4].
Table 1: Impact of Experimental Parameters on Bacterial Recovery from Forest Soil Using a Diffusion Bioreactor [4]
| Parameter | Condition Tested | Key Finding on Recovery |
|---|---|---|
| Cultivation Technique | Novel Diffusion Bioreactor | Successfully cultivated 35 previously uncultured strains. |
| Traditional Shake Flask/Plating | No previously uncultured strains were recovered. | |
| Sampling Season | Summer (e.g., July, August) | Increased recovery of uncultured and novel isolates. |
| Winter (e.g., February, January) | Lower recovery of novel diversity. | |
| Incubation Period | Prolonged (up to 4 weeks) | Critical for the recovery of slow-growing species. |
| Culture Media | Low-substrate (e.g., 50% R2A, R2A-SE) | Enhanced recovery of novel and uncultured isolates. |
| Nutrient-rich (e.g., TSA, LB) | Favored fast-growing, commonly cultured species. |
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Materials and Reagents for In-Situ Cultivation Experiments
| Item | Function/Application | Specific Examples & Notes |
|---|---|---|
| Semi-Permeable Membrane | Forms a physical barrier that permits diffusion of molecules but contains cells. | Polycarbonate membrane (0.4 µm pore size) [4]. |
| Low-Gelling Temperature Agarose | Used for microencapsulation of cells; gels at low temperatures to maintain cell viability [31]. | Sigma A9414, prepared in diluted culture medium at 35°C [31]. |
| Dialysis Cassette | Serves as a ready-made, robust diffusion chamber for in-situ incubation. | Modified Slyde-A-Lyzer cassettes [31]. |
| Culture Media | Provides basal nutrients for growth. Low-nutrient media are often superior. | R2A, 50% Diluted R2A, Soil Extract (SE)-supplemented media [4]. |
| Soil Extract (SE) | Adds habitat-specific nutrients and trace elements that may be critical for growth of fastidious organisms [4]. | Prepared by centrifuging and filter-sterilizing an aqueous soil slurry. |
| Cycloheximide | An antifungal agent to suppress fungal contamination in bacterial cultures. | Used at 50 µg/mL in culture media [4]. |
| N-Boc-Tris | N-Boc-Tris, CAS:146651-71-0, MF:C9H19NO5, MW:221.25 g/mol | Chemical Reagent |
| Triammonium phosphate trihydrate | Triammonium phosphate trihydrate, CAS:25447-33-0, MF:H18N3O7P, MW:203.13 g/mol | Chemical Reagent |
Technique Selection and Workflow
Choosing the right technique and understanding the microbial responses to their environment are crucial for success. The following diagram illustrates the decision-making process for technique selection and the biological principles underlying in-situ cultivation.
Troubleshooting Guides and FAQs
Frequently Asked Questions (FAQs)
Q1: Why is the population ratio in my synthetic consortium unstable over time? The instability often arises from competitive or antagonistic relationships between member species, where faster-growing microbes outcompete others for limited resources like phosphate and nitrogen, even in nutrient-rich environments [34] [35]. This "winner-takes-all" dynamic can collapse the intended community structure. To mitigate this, design cross-feeding dependencies or allocate different carbon sources to different species to reduce direct competition [35].
Q2: How can I determine the interaction types between species in my consortium? Computational modeling tools like the generalized Lotka-Volterra (gLV) model can predict interaction types and their impact on community diversity [34] [36]. Furthermore, Dynamic Flux Balance Analysis (dFBA) can model temporal metabolic exchanges and emergent interactions during community assembly [34]. These tools provide mechanistic insights into the ecological relationships within your consortium.
Q3: What should I do if my coculture shows reduced overall biomass or product yield? Reduced yield can result from metabolic imbalances or competitive interactions that suppress the growth of individual members compared to their monoculture performance [34] [35]. Establish mutualistic cross-feeding interactions, for example, by engineering co-dependence on exchanged metabolites like amino acids or vitamins, to shift the relationship toward symbiosis and enhance system stability and output [35].
Q4: How does the initial inoculation ratio affect the final community structure? Research on a model three-member rhizosphere community found that different initial inoculum ratios (varying by up to three orders of magnitude) did not significantly alter the final community structure or the competitive interaction patterns [34]. The community consistently reached the same stable composition, suggesting that in some systems, emergent interactions rather than initial conditions dictate the final outcome.
Troubleshooting Common Coculture Problems
Problem: Rapid Overgrowth by One consortium Member
- Potential Causes: Direct competition for a primary nutrient source (e.g., glucose, phosphate); lack of growth-inhibiting interactions toward the dominant member [35].
- Solutions:
- Resource Partitioning: Genetically engineer members to utilize different carbon substrates. For example, knock out glucose transporter genes in one strain to force the use of another carbon source like xylose [35].
- Establish Cross-Feeding: Design the system so that the overgrown member relies on a metabolite provided by another member, creating a mutualistic check on its population [35].
Problem: Low Product Titer Despite Good Cell Growth
- Potential Causes: Metabolic burden; inefficient metabolite transfer between members leading to dilution in the extracellular environment [35].
- Solutions:
- Spatial Engineering: Use materials or biofilms to spatially organize the members, potentially reducing diffusion distances and improving metabolic exchange efficiency [37].
- Pathway Optimization: Rebalance the metabolic flux by adjusting gene expression levels in the different members to minimize the accumulation of inhibitory intermediates [35].
Problem: Consortium Fails to Stabilize After Serial Transfer
- Potential Causes: Antagonistic interactions, such as one member producing inhibitory compounds; lack of essential cross-feeding metabolites [35].
- Solutions:
- Screen for Compatibility: Before final assembly, test all pairwise interactions to identify and replace strains that exhibit strong antagonism [35].
- Adaptive Laboratory Evolution (ALE): Coculture the members over multiple generations to allow for the evolution of stable, cooperative interactions, as demonstrated in systems where vitamin-producers were evolved to support vitamin-dependent yeast [35].
Summarized Experimental Data
Table 1: Population Dynamics in a Model Three-Member Bacterial Community in Nutrient-Rich Medium [34]
| Bacterial Strain | Relative Abundance in Stabilized Community (Plate Count) | Relative Abundance in Stabilized Community (qPCR) | Growth in Coculture vs. Monoculture |
|---|---|---|---|
| Pseudomonas sp. GM17 | Dominant (Majority) | Dominant (Majority) | Significantly decreased |
| Pantoea sp. YR343 | Intermediate | Higher than plate count | No significant difference (plate count) / Decreased (qPCR) |
| Sphingobium sp. AP49 | Low | Low | Decreased by ~two orders of magnitude |
Table 2: Computational Tools for Modeling and Designing Microbial Consortia [34] [36]
| Tool Name | Type | Primary Function | Application Example |
|---|---|---|---|
| Generalized Lotka-Volterra (gLV) | Ecological Model | Predicts interaction types (e.g., competition, cooperation) between species and their effect on community diversity. | Analyzing succession and stability in a three-strain rhizosphere community [34]. |
| Dynamic Flux Balance Analysis (dFBA) | Metabolic Model | Models time-dependent microbial growth and metabolite-mediated interactions using genome-scale metabolic models. | Predicting temporal metabolic exchanges during community assembly [34]. |
| COMETS (Computation of Microbial Ecosystems in Time and Space) | Software Platform (extends dFBA) | Simulates spatiotemporal dynamics and metabolic interactions of microbial communities. | Providing mechanistic insight into community structure emergence [34]. |
| Treg Induction Score (TrIS) | Computational Index | Scores and ranks microbial consortia by their predicted potential to induce a specific immune response (e.g., Treg cells). | Selecting optimal Clostridia consortia for immune modulation in mice [36]. |
Detailed Experimental Protocols
Protocol 1: Constructing and Tracking a Synthetic Microbial Community
This protocol is adapted from studies investigating the formation of stabilized communities in nutrient-rich conditions [34].
1.1 Bacterial Strains and Medium
- Strains: Pseudomonas sp. GM17, Pantoea sp. YR343, and Sphingobium sp. AP49 (or your chosen isolates).
- Medium: R2A complex medium (or another nutrient-rich medium relevant to the isolation environment).
1.2 Inoculation and Serial Transfer
- Prepare the initial co-culture by inoculating the three strains in R2A medium at the desired initial ratio (e.g., 1:1:1, or varied for testing priority effects).
- Incubate at an appropriate temperature (e.g., 30°C) with shaking for 24 hours (or one growth cycle).
- After the growth cycle, serially transfer the community by diluting an aliquot (e.g., 1:100) into fresh R2A medium.
- Repeat this transfer for multiple cycles (e.g., 5-10 cycles) to allow the community to stabilize.
1.3 Tracking Community Structure
- Plate Counting: At each transfer time point, serially dilute the culture and spread plate on solid R2A agar. Identify and count the different species based on colony morphology.
- Quantitative PCR (qPCR): As a complementary and often more precise method, extract total DNA from the community at each time point. Perform qPCR using strain-specific primers to quantify the abundance of each member. Calculate the relative percentage of each species.
1.4 Data Analysis
- Plot the relative abundance of each species over time for both plate counting and qPCR data to visualize community succession and stabilization.
- Compare the final abundance of each species in coculture to its maximum abundance in monoculture to infer interaction types (e.g., competition if growth is decreased).
Protocol 2: Computational Simulation of Community Dynamics using gLV
This protocol outlines how to use the gLV model to predict microbial interactions [34] [36].
2.1 Model Formulation The generalized Lotka-Volterra model for multiple species is defined by the equation:
( \frac{dXi}{dt} = \mui Xi \left(1 - \frac{\sum{j=1}^n \alpha{ij} Xj}{K_i}\right) )
Where:
- ( X_i ) is the biomass (or abundance) of species ( i ).
- ( \mu_i ) is the intrinsic growth rate of species ( i ).
- ( K_i ) is the carrying capacity of species ( i ).
- ( \alpha_{ij} ) is the interaction coefficient, representing the effect of species ( j ) on species ( i ).
2.2 Parameterization
- Monoculture Data: Grow each species in monoculture to estimate its intrinsic growth rate (( \mui )) and carrying capacity (( Ki )).
- Interaction Terms (( \alpha_{ij} )): Estimate the interaction coefficients by fitting the gLV model to time-series data of all species in coculture. This can be done using nonlinear regression or other model-fitting algorithms.
2.3 Simulation and Prediction
- Use the parameterized gLV model to simulate the population dynamics of the community over time from different starting conditions.
- Validate the model by comparing its predictions with experimental data not used in the fitting process (e.g., from different initial inoculum ratios).
Visualized Workflows and Relationships
Coculture Development Workflow
Microbial Interaction Relationships
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Tools for Microbial Consortium Research
| Item | Function/Benefit | Example Application |
|---|---|---|
| R2A Complex Medium | A nutrient-rich medium suitable for cultivating a wide range of environmental bacteria, mimicking resource-abundant environments like the rhizosphere [34]. | Serving as the growth medium to study community assembly without obligate metabolic dependencies [34]. |
| Strain-Specific qPCR Primers | Enable precise quantification of individual species' abundance within a mixed community, bypassing limitations of morphology-based colony counting [34]. | Tracking the population dynamics of each consortium member over serial transfer cycles [34]. |
| COMETS Software Platform | A powerful bioinformatics tool that performs dFBA simulations to predict spatiotemporal metabolic interactions in microbial communities [34]. | Modeling and predicting metabolite exchanges and emergent interactions during consortium stabilization [34]. |
| Mycoplasma Detection Kit | Critical for routine quality control to detect mycoplasma contamination, which can alter cell metabolism and gene expression without obvious visual signs [38] [39]. | Ensuring that mammalian cell cocultures or host-interaction experiments are not compromised by underlying contamination [40]. |
| Antibiotics for Selection | Used to maintain plasmid stability or select for specific engineered strains within a consortium. Use with caution to avoid inducing gene expression changes [38]. | Maintaining a desired population ratio in a consortium of engineered strains, each with different antibiotic resistance markers. |
| 9-Fluorenylmethanol | 9-Fluorenemethanol, 99%|CAS 24324-17-2|RUO | 9-Fluorenemethanol, a key synthetic intermediate for Fmoc peptide chemistry. ≥98% purity. For Research Use Only. Not for human or veterinary use. |
This technical support center provides practical guidance for researchers tackling the challenge of cultivating previously uncultured microorganisms. The following troubleshooting guides and FAQs address common experimental hurdles in using genomic data to design custom growth media, supporting broader efforts in microbial cultivation for drug discovery and environmental research.
Troubleshooting Guide: Common Targeted Isolation Challenges
Problem 1: Poor or No Growth Despite Genomic Evidence of Metabolic Potential
Question: My microorganism's genome suggests all necessary metabolic pathways are present, but I'm not observing growth in my designed medium. What could be wrong?
Answer: This common issue often stems from mismatches between genomic potential and actual cultivation requirements. Try these solutions:
- Verify nutrient concentrations: Genomic evidence often indicates trophic strategy. For genome-streamlined oligotrophs, reduce carbon content to 1-2 mg DOC per liter, as high nutrient levels can inhibit growth [10].
- Check for auxotrophies: Look for missing biosynthetic pathways in the genome that suggest dependencies on specific growth factors. Supplement with yeast extract (0.001-0.01%) or a vitamin mixture to cover potential gaps [10].
- Recreate environmental conditions: If the source environment data is available, match pH, temperature, and ionic composition. For freshwater oligotrophs, successful cultivation often occurs at 16°C with μM concentrations of organic compounds [10].
- Extended incubation: Slowly growing oligotrophs may require 6-8 weeks before visible growth appears [10].
Problem 2: Contamination Outcompetes Target Organism
Question: My custom media supports growth, but fast-growing contaminants overwhelm my target microorganism. How can I address this?
Answer: This typically occurs when media conditions favor copiotrophs over your target organism:
- Apply dilution-to-extinction: Inoculate multiple wells with approximately one cell per well to separate target organisms from competitors [10].
- Reduce nutrient levels: Switch to low-nutrient defined media that specifically disadvantages fast-growing copiotrophs [10].
- Use semi-permeable membranes: Cultivate cells in diffusion chambers that allow nutrient exchange while providing physical separation [1].
- Add resuscitation factors: Supplement with Micrococcus luteus culture supernatant containing resuscitation-promoting factor (Rpf) to stimulate dormant cells [6].
Problem 3: Growth Initiates but Cannot Be Sustained
Question: I observe initial growth in primary cultivation, but the culture cannot be maintained through transfers. What might be causing this?
Answer: This suggests missing factors in your transfer media:
- Identify essential cofactors: Genomic analysis may reveal dependencies on specific metabolites. Test additions of catalase, various vitamins, or specific carbon sources [10].
- Check for metabolic byproduct dependencies: Some microorganisms require metabolites produced by other bacteria. Consider co-culture with suspected partner organisms [1].
- Verify energy sources: Ensure your medium contains the specific energy sources (organic acids, carbohydrates, methanol) indicated by genomic analysis of respiratory pathways [10].
Frequently Asked Questions
Q1: What genomic features most reliably inform media design? Focus on identifying complete metabolic pathways rather than individual genes. Key elements include: central carbon metabolism pathways, energy generation systems, biosynthetic capabilities for essential metabolites, and apparent auxotrophies (missing pathways for essential compounds) [41] [10].
Q2: How can I determine optimal nutrient concentrations from genomic data? Genome size and coding density often correlate with trophic strategy. Small, streamlined genomes (<2 Mbp) with high coding density typically indicate oligotrophs requiring 1-10 mg carbon/L. Larger genomes with more regulatory genes often suggest copiotrophs tolerating richer media (>1 g carbon/L) [10].
Q3: What if the genome suggests numerous potential carbon sources? Begin with defined media containing 5-10 different carbon sources at μM concentrations, focusing on compounds present in the native habitat. Test mixtures of carbohydrates, organic acids, and amino acids initially, then refine based on growth response [10].
Q4: How long should I incubate cultures before declaring failure? Standard incubation periods may be insufficient. While clinical isolates often grow in 24-48 hours, environmental oligotrophs may require 6-8 weeks. Monitor for growth monthly for at least 3 months before concluding the experiment failed [10] [6].
Q5: Can machine learning help predict growth requirements from genomic data? Yes, emerging approaches show promise. Models using 16S rRNA sequence features (like 3-mer frequencies) can predict media compatibility with 76-99.3% accuracy, providing useful starting points for media design [42].
Experimental Protocols & Data
Protocol 1: High-Throughput Dilution-to-Extinction Cultivation
This method effectively isolates oligotrophic microorganisms while minimizing competition from fast-growing copiotrophs [10]:
- Sample preparation: Filter environmental sample through 0.2 μm membrane, resuspend cells in sterile basal salts solution.
- Media preparation: Prepare defined artificial media mimicking natural conditions (see Table 1 for composition).
- Inoculation: Dilute cell suspension to approximately 1 cell/well and dispense into 96-deep-well plates containing media.
- Incubation: Maintain at temperature matching source environment (e.g., 16°C for freshwater lakes) for 6-8 weeks.
- Growth screening: Monitor turbidity or DNA staining weekly. Transfer positive wells to fresh media.
- Purity verification: Confirm axenic status by 16S rRNA gene sequencing and repeated streaking.
Protocol 2: Genomically-Informed Media Design Workflow
Systematic approach for translating genomic data into cultivation media [41] [10]:
- Genome analysis: Annotate genome using RAST, IMG/M, or similar platform.
- Metabolic reconstruction: Identify complete pathways for energy generation, carbon utilization, and biomass synthesis.
- Auxotrophy detection: Note missing pathways for essential metabolites (amino acids, vitamins, cofactors).
- Environmental matching: Incorporate geochemical data from sampling environment when available.
- Media formulation: Design defined medium containing:
- Energy sources and electron acceptors identified from genome
- Essential salts and minerals
- Required growth factors indicated by auxotrophies
- Potential signaling compounds (if suggested by genome)
- Iterative refinement: Test multiple versions with variations in component concentrations.
| Component | Medium 2 (mg/L) | Medium 3 (mg/L) | MM-Medium (mg/L) | Function |
|---|---|---|---|---|
| Carbon Sources | ||||
| Glucose | 0.5 | - | - | Carbon/energy |
| Fructose | 0.5 | - | - | Carbon/energy |
| Succinate | 0.5 | 0.5 | - | Carbon/energy |
| Acetate | 0.5 | 0.5 | - | Carbon/energy |
| Methanol | - | - | 100 | C1 carbon |
| Methylamine | - | - | 100 | C1 carbon |
| Vitamins | ||||
| Cyanocobalamin (B12) | 0.0005 | 0.0005 | 0.0005 | Cofactor |
| Thiamine (B1) | 0.1 | 0.1 | 0.1 | Cofactor |
| Biotin | 0.005 | 0.005 | 0.005 | Cofactor |
| Other | ||||
| Catalase | 0.1 | 0.1 | 0.1 | Oxidative protection |
| MOPS buffer | 100 | 100 | 100 | pH stabilization |
| Trace elements | 1 mL | 1 mL | 1 mL | Micronutrients |
Table 2: Troubleshooting Common Cultivation Failure Scenarios
| Problem | Genomic Indicators | Potential Solutions | Success Metrics |
|---|---|---|---|
| No growth | Complete central metabolism but missing cofactor synthesis | Supplement with 0.001% yeast extract or vitamin mix | Visible turbidity within 4-8 weeks |
| Growth then death | Evidence of oxidative stress sensitivity | Add 0.1-1 mg/L catalase or 0.1% pyruvate | Sustained growth through >3 transfers |
| Poor growth yield | Streamlined genome, transporter-rich | Reduce carbon to 1-2 mg/L DOC, use multiple carbon sources | Increased cell density (>10^7 cells/mL) |
| Contamination | No specific genomic indicators | Apply dilution-to-extinction, use selective antibiotics | Axenic culture verified by sequencing |
Research Reagent Solutions
Essential Materials for Targeted Isolation Experiments
| Reagent/Category | Specific Examples | Function in Cultivation |
|---|---|---|
| Basal Salt Mixtures | Artificial freshwater medium, Marine salts mixture | Provides essential ions and osmotic balance matching natural habitat |
| Carbon Sources | Carbohydrate mixtures, Organic acids, Methanol/methylamine | Energy and carbon skeletons based on genomic predictions |
| Growth Factors | Vitamin mixtures, Yeast extract, Resuscitation-promoting factors (Rpf) | Supplements auxotrophies and stimulates dormant cells |
| Gelling Agents | Gellan gum, Agarose | Solid support for isolation while allowing nutrient diffusion |
| Detection Reagents | SYBR Gold, CTC dye, Radioactive substrates | Visualizing microbial growth when turbidity is minimal |
| Cultivation Devices | Diffusion chambers, Microtiter plates, Cell sorting equipment | Physical separation and high-throughput processing |
Advanced Methodologies
Genome-Based Media Optimization Framework
Recent advances enable more systematic approaches to cultivating previously uncultured taxa:
Machine Learning Applications: Binary classification models using 16S rRNA sequence features (3-mer frequencies) can predict microbial growth on specific media with 76-99.3% accuracy, guiding initial media selection [42].
Metagenome-Informed Cultivation: When pure genome sequences are unavailable, metagenome-assembled genomes (MAGs) from the same environment can provide metabolic insights for designing targeted media [10].
High-Throughput Screening: Testing numerous media variations in parallel using 96-well formats significantly increases success rates. One study achieved 40% recovery of previously uncultured freshwater taxa using this approach [10].
For further assistance with specific cultivation challenges, consult the centralized culture collection at http://isolates.reefgenomics.org/ or reference the genomic resources cited in this guide [41].
FAQs: Addressing Common High-Throughput Workflow Challenges
FAQ 1: What are the primary challenges in cultivating unculturable gut microbial species, and how can high-throughput workflows address them? The primary challenge is that a significant proportion of gut microbiota cannot grow using traditional plate-based techniques, a phenomenon known as the "great plate count anomaly," often because the culture media do not accurately represent the natural gut environment [43]. High-throughput culturomics addresses this by employing a vast array of culture conditions (e.g., 212 different conditions) and nutrient substrates like blood culture bottles, rumen fluid, and sheep blood to optimize for species diversity [43]. These methods have successfully expanded the repertoire of cultivable human gut species [43].
FAQ 2: How can I improve the recovery of complete microbial genomes, especially the largest gene segments, from samples with low viral or bacterial load? Recovery of large gene segments from low-biomass samples can be challenging due to inefficient amplification [44]. An optimized multisegment RT-PCR (mRT-PCR) protocol can enhance the amplification of all genomic segments. This involves using a specific RT enzyme (e.g., LunaScript RT Master Mix) and adapting the RT and PCR cycling conditions, which has been shown to improve sensitivity and overall recovery of all genomic segments compared to established protocols [44].
FAQ 3: What strategies can be implemented to make high-throughput whole-genome sequencing more cost-effective for public health laboratories? Implementing a streamlined, cost-effective laboratory workflow is key. Specific strategies include [45]:
- Using automated high-throughput DNA extraction systems (e.g., QIAcube HT).
- Adopting quarter-volume reactions for sequencing library preparation (e.g., Illumina DNA Prep with quarter volumes).
- Utilizing efficient sequencing platforms (e.g., NextSeq). This combined approach allows for processing a high volume of samples while reducing reagent costs and maintaining a median turnaround time of 7 days [45].
FAQ 4: How can workflow automation and informatics tools benefit a high-throughput screening lab? Workflow automation offers several critical benefits for screening labs [46]:
- Increased Productivity: Automating repetitive tasks allows scientists to focus on complex aspects of high-throughput biology.
- Enhanced Data Integrity: Standardized workflows and automated data capture improve the accuracy and reliability of data.
- Accelerated Discovery: Efficient workflow management expedites the screening process, leading to faster identification of therapeutic candidates.
- Resource Optimization: Effective workflow design helps allocate resources judiciously, minimizing waste of reagents and in vitro systems.
FAQ 5: What are the best practices for managing the large amounts of data generated by high-throughput workflows? Best practices involve strategic workflow management [46]:
- Process Standardization: Develop and enforce standard operating procedures for consistency across screening platforms.
- Data Flow Management: Implement informatics systems to seamlessly capture, store, and track experimental data from inception to analysis.
- Continuous Improvement: Regularly monitor workflow performance to identify bottlenecks and areas for optimization.
Troubleshooting Guides
Problem 1: Low Bacterial Diversity in Cultured Isolates
Potential Causes and Solutions:
| Cause | Solution | Reference |
|---|---|---|
| Insufficient variety in culture conditions. | Implement a culturomics approach. Use a wide array of culture media (dozens to hundreds) with different nutrient substrates, such as rumen fluid and sheep blood, to mimic various gut niches. | [43] |
| Overgrowth by fast-growing species. | Use droplet microfluidics. Technologies like the microfluidic streak plate or SlipChip physically separate individual bacterial cells in nanoliter-to-picoliter droplets, preventing competition and allowing slow-growing species to thrive. | [43] |
| Oxygen sensitivity of anaerobic species. | Integrate the entire cultivation workflow, including droplet generation and incubation, within an anaerobic glove box to protect extremely oxygen-sensitive (EOS) strains. | [47] [43] |
Problem 2: Poor Yield or Incomplete Genome Assembly from Sequencing
Potential Causes and Solutions:
| Cause | Solution | Reference |
|---|---|---|
| Inefficient amplification of all genomic segments. | Optimize your RT-PCR protocol. Use a high-performance RT enzyme and tailor the RT and PCR cycling conditions to improve the sensitivity of recovering whole-genome sequences, especially for the largest polymerase genes. | [44] |
| Sequencing library preparation is not cost-effective for high volume. | Scale down reaction volumes. For Illumina library prep, using a quarter-volume reaction can maintain data quality while significantly reducing per-sample costs, enabling higher throughput. | [45] |
| Choosing the wrong sequencing technology for the application. | Select technology based on project goals. For high accuracy and throughput, use Illumina. For long reads to resolve complex regions, use PacBio or Oxford Nanopore. | [48] |
Problem 3: Workflow Bottlenecks and Inefficiencies
Potential Causes and Solutions:
| Cause | Solution | Reference |
|---|---|---|
| Manual, repetitive tasks are slowing down the workflow. | Integrate automation and workflow management tools. Use automated colony pickers, liquid handlers, and software like KanBo or Snakemake to standardize processes, track tasks, and automate data flow. | [43] [46] |
| Input/Output (IO) operations are causing system slowdowns. | Optimize data handling. For workflows involving thousands of small files, use the fast local NVMe disk on compute nodes or containerize applications with SquashFS to reduce excessive load on the shared file system (Lustre). | [49] |
| Lack of clear documentation leads to confusion and errors. | Create and use standardized templates. Develop SOPs and template protocols for repetitive tasks like assay development or data analysis to ensure consistency and reduce errors across the team. | [50] [46] |
Workflow Visualization
Integrated HTS Cultivation-to-Sequencing Workflow
Troubleshooting Decision Pathway
Essential Research Reagent Solutions
Cultivation and Isolation Reagents
| Reagent / Solution | Function in the Workflow |
|---|---|
| Diverse Culture Media | Used in culturomics to emulate various gut nutritional niches, enabling the growth of fastidious bacteria [43]. |
| Blood Culture Bottles | Serves as a critical nutrient-rich substrate for cultivating a wider range of gut microbes [43]. |
| AnaeroPack / GasPak Systems | Chemically generates an anaerobic environment essential for cultivating oxygen-sensitive gut anaerobes [43]. |
| Microfluidic Oil & Cartridges | Used in droplet microfluidic systems to generate millions of picolitre droplets for single-cell isolation and cultivation [43]. |
Sequencing and Analysis Reagents
| Reagent / Solution | Function in the Workflow |
|---|---|
| LunaScript RT Master Mix | A reverse transcription enzyme mix used in optimized protocols for enhanced cDNA synthesis from viral RNA [44]. |
| Q5 Hot Start High-Fidelity DNA Polymerase | Used in the PCR amplification step post-reverse transcription to ensure accurate amplification of genomic segments [44]. |
| Illumina DNA Prep Kit | A library preparation kit for whole-genome sequencing. Using quarter-volume reactions makes it cost-effective for high-throughput [45]. |
| Oxford Nanopore Ligation Sequencing Kit | Used for library preparation on Nanopore platforms, enabling real-time, long-read sequencing [48]. |
| AMPure XP Beads | Used for PCR amplicon clean-up and size selection to remove primers and fragments smaller than 500 bp before sequencing [44]. |
Optimizing for Success: Strategies to Overcome Growth and Isolation Hurdles
Why is Media Formulation Critical for Oligotrophs? Oligotrophic microorganisms are adapted to nutrient-scarce environments. Standard laboratory media, which are often nutrient-rich, can inhibit their growth due to osmotic stress, the promotion of fast-growing "weed" species, or the failure to provide essential trace nutrients found in their natural habitats [1] [51] [28]. Successfully cultivating these organisms requires a deliberate shift in strategy from providing nutrient excess to carefully balancing nutrient concentration and stoichiometry to mimic their native conditions.
A key concept in this balance is stoichiometric plasticity, which refers to the ability of an organism or community to vary its elemental composition (e.g., C:N:P ratios) in response to environmental nutrient availability [52]. For instance, in marine environments, the community phosphorus-to-carbon ratio (P:C) exhibits a linear relationship with ambient phosphate concentrations, while the nitrogen-to-carbon ratio (N:C) is more rigid and only responds at very low nitrate levels [52]. This greater plasticity in P requirements is a fundamental principle to consider when formulating media.
Troubleshooting Guides
Common Cultivation Failure Modes and Solutions
Problem: No growth observed in initial cultures.
- Potential Cause 1: Medium is too rich, causing toxicity or favoring competitors.
- Solution: Dilute the standard medium by 10 to 100-fold with sterile water or a defined salts solution. Alternatively, use a minimal defined medium with a low carbon concentration (µg/L to mg/L range) [51].
- Potential Cause 2: Essential growth factors from the natural environment are missing.
- Solution: Supplement the medium with filtered (0.22 µm) water or soil extract from the source environment. Alternatively, use dissolved organic matter (DOM) extracted from the native habitat as a complex carbon source [28].
- Potential Cause 3: Target cells are entering a Viable But Non-Culturable (VBNC) state.
Problem: Initial growth ceases upon subculturing to fresh media.
- Potential Cause 1: The organism depends on metabolic byproducts or signaling molecules from other community members (co-dependency).
- Potential Cause 2: Accumulation of inhibitory compounds from agar preparation.
- Solution: Autoclave agar and phosphate buffers separately to prevent the formation of growth-inhibiting hydrogen peroxide [51].
Problem: Unintended organisms (contaminants/weeds) overgrow the culture.
- Potential Cause: The nutrient profile favors fast-growing generalists (r-strategists).
- Solution: Use a carbon source that is more recalcitrant or specific, such as lignin-derived compounds from sediment DOM or specific aromatic acids. This selectively enriches for K-strategists (slow-growing, oligotrophic specialists) [28].
Advanced Troubleshooting: Community Interaction Co-Limitation (CIC)
In a microbial community, the limitation of one organism can cause the limitation of another through metabolic dependencies, a phenomenon termed Community Interaction Co-Limitation (CIC) [53]. For example, if vitamin B12-producing bacteria are limited by iron, the phytoplankton that depend on that vitamin will also become co-limited, even if the primary macronutrients (N, P) are available [53].
- Symptoms: Failure of a community to grow despite supplementation with N, P, and C, or a collapse of a previously stable co-culture.
- Diagnosis Strategy: Systematically supplement with potential "biologically produced nutrients" (BPNs) like vitamins (B12, B1) or specific amino acids, in addition to major nutrients [53].
- Solution: Identify and relieve the nutrient limitation of the "helper" population (e.g., the vitamin producer) to restore the function of the dependent population.
Frequently Asked Questions (FAQs)
Q1: What is the single most important adjustment when switching from culturing model organisms to oligotrophs? The most critical adjustment is reducing the concentration of carbon and energy sources. Oligotrophs are adapted to low nutrient flux, and standard media concentrations can be toxic or selectively inhibit their growth [51] [28].
Q2: How do I determine the optimal N:P ratio for my target organism's medium? There is no universal optimal ratio, as it depends on the organism's innate stoichiometry and plasticity. A successful strategy is to start with the Redfield ratio (C:N:P = 106:16:1) and then adjust based on environmental data. If the source environment is known to have a high N:P ratio (>100:1), formulate a medium with a similarly high ratio to selectively enrich for organisms adapted to phosphorus scarcity [52] [54].
Q3: Can I use complex carbon sources like yeast extract? While yeast extract can be useful, it is a poorly defined mixture that primarily promotes fast-growing bacteria. For greater success with previously uncultured taxa, consider using more environmentally relevant complex carbon sources, such as sediment-derived dissolved organic matter (DOM) or bacterial cell lysate, which provide a more natural mixture of labile and recalcitrant compounds [28].
Q4: What operational bioreactor strategies are best for nutrient-limited growth? The chemostat is the ideal tool for studying nutrient-limited growth, as it allows precise control over the growth rate and the limiting nutrient. For bioproduction, fed-batch processes where the feed rate is carefully controlled to maintain nutrient limitation can prevent the buildup of inhibitory by-products and help sustain stable growth phases that are not possible in batch culture [55].
Q5: Why would adding other living bacteria help my target bacterium grow? Many unculturable bacteria exist in complex metabolic networks. Co-culture with "helper" bacteria can provide essential growth factors, remove inhibitory waste products, or modify the chemical environment in a beneficial way that cannot be replicated with a defined medium alone [1] [51].
Quantitative Data & Stoichiometry Tables
Table 1: Stoichiometric Plasticity in Marine Phytoplankton Community
Table based on a large-scale analysis of particulate matter, showing how community nutrient ratios respond to ambient concentrations [52].
| Ambient Nutrient Concentration | Nitrogen-to-Carbon (N:C) Response | Phosphorus-to-Carbon (P:C) Response |
|---|---|---|
| Low NO₃ (0-3 µM) | Increases by ~20% | Not Applicable |
| High NO₃ (>3 µM) | Saturates; becomes constant | Not Applicable |
| Low POâ‚„ | Not Applicable | Increases linearly with POâ‚„ |
| Empirical Relationship | N:C saturates at high NO₃ | P:C (‰) = 6.9 ‰ µMâ»Â¹ × [POâ‚„] + 6.0 ‰ |
Table 2: Saturation Constants (Kâ‚›) for E. coli with Glucose
This table illustrates the low substrate concentrations at which growth becomes nutrient-limited, highlighting the need for precise feeding strategies [55].
| Strain / Condition | Saturation Constant (Kâ‚›) for Glucose | Reference Context |
|---|---|---|
| Low-end affinity | 50 µg/L (≈ 0.28 µM) | [55] |
| High-end value | > 8 mg/L (≈ 44 µM) | [55] |
| Typical Batch Culture | 1-10 g/L (>> Kâ‚›) | Growth is always at µₘâ‚â‚“ until depletion |
Experimental Protocols & Workflows
Protocol 1: Preparation of Sediment Dissolved Organic Matter (DOM) for Media
Objective: To extract a natural, complex carbon source from an environmental sample to enhance the cultivation of oligotrophic subsurface bacteria [28].
Materials:
- Sediment from the target environment (e.g., groundwater well)
- Milli-Q water
- Centrifuge and bottles
- Rotary shaker
- Sonicator (water bath)
- Freeze dryer (lyophilizer)
- 0.22 µm PES membrane filtration system
Method:
- Extraction: Combine freeze-dried sediment with Milli-Q water at a 1:4 (w/w) ratio. Shake the mixture on a rotary shaker (170 rpm) overnight at 35°C.
- Sonication: Sonicate the water-sediment slurry in a water bath for 2 hours.
- Separation: Centrifuge the slurry at 6,000 g for 20 minutes. Carefully decant and collect the supernatant.
- Sterilization: Filter the supernatant through a 0.22 µm PES membrane filter.
- Concentration & Storage: Freeze-dry the filtrate to obtain a solid DOM material. Store the lyophilized DOM at -20°C until use. For media preparation, dissolve the DOM in filtered, sterile water to the desired concentration (e.g., 200 mg/L) [28].
Protocol 2: Dilution-to-Extinction Cultivation in a Simulated Natural Environment
Objective: To isolate slow-growing, oligotrophic bacteria by drastically reducing competition and exposing cells to a continuous flux of natural growth factors [1] [51].
Materials:
- Dilute suspension of cells from the environmental sample (e.g., water, soil)
- Dilute nutrient broth (e.g., 1/100 R2A broth) or a very minimal defined medium
- Diffusion chambers (e.g., semi-permeable membranes mounted on a ring) or commercial culture capsules
- Aquarium or container to hold natural water and sediment from the sample site
Method:
- Inoculate: Place the diluted cell suspension inside the diffusion chamber.
- Incubate In Situ: Seal the chamber and incubate it within an aquarium containing natural water and a bed of sediment from the source environment. This allows chemical exchange with the natural habitat.
- Monitor: Observe the chambers microscopically for the formation of microcolonies, which may take weeks.
- Isolate: Once microcolonies are established, open the chamber and subculture onto solid media prepared with the same dilute nutrients and environmental extracts [1].
Key Experimental Workflow Diagram
Diagram Title: Oligotroph Cultivation Troubleshooting Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Oligotroph Media Formulation
| Reagent / Material | Function & Rationale | Key Considerations |
|---|---|---|
| Sediment DOM [28] | A complex, natural carbon source that provides a spectrum of labile and recalcitrant compounds, mimicking the in-situ nutrient profile. | More effective than simple C sources for cultivating diverse, distinct, and novel bacteria. |
| Bacterial Cell Lysate [28] | Provides a complex mixture of nutrients, including cellular components and metabolites, which can serve as growth factors for dependent organisms. | Use a strain from the same environment to increase ecological relevance. |
| Semi-Permeable Membranes (for diffusion chambers) [1] [51] | Allows the passage of nutrients and growth factors from the natural environment while containing the target cells, simulating their natural habitat. | Crucial for cultivating organisms with unknown growth factor requirements. |
| Environmental Extracts (Soil/Water) [51] | A source of undefined but environmentally relevant trace metals, vitamins, and co-factors that may be absent in synthetic media. | Should be filter-sterilized (0.22 µm) before addition to media. |
| Defined Trace Metal Mixes | To investigate specific micronutrient limitations (e.g., Fe, Co, Zn) and formulate stoichiometrically balanced media. | Essential for testing hypotheses related to co-limitation (e.g., Fe and B12) [53]. |
| Recalcitrant Carbon Sources (e.g., lignin, aromatics) | Selectively enriches for slow-growing oligotrophs (K-strategists) capable of degrading complex compounds, reducing competition from fast-growing weeds. | Can be used in combination with labile C sources in very low concentrations. |
In the pursuit of cataloging Earth's microbial diversity, researchers face a significant challenge: the majority of microorganisms resist cultivation in laboratory settings. This problem stems largely from the intrinsic competition between microbial life strategies, where fast-growing copiotrophs rapidly consume standard laboratory nutrients, overwhelming their slower-growing oligotrophic counterparts. This technical guide explores advanced methodologies to overcome this competition barrier, enabling the isolation and study of previously "unculturable" species with unique physiological adaptations and biotechnological potential.
FAQs & Troubleshooting Guides
FAQ 1: What are the primary reasons slow-growing microbes are outcompeted in standard culturing?
Answer: Slow-growing microbes, often oligotrophs adapted to nutrient-scarce environments, face several disadvantages in standard rich media:
- Growth Rate Disparity: Fast-growing copiotrophs exhibit significantly higher maximal growth rates (µMax), quickly dominating shared nutrient pools [56].
- Resource Competition: In batch culture systems, copiotrophs rapidly deplete essential nutrients before slow-growing species can establish substantial populations [56] [1].
- Inhibitory Byproducts: Metabolic activity of fast-growing species often produces antimicrobial compounds or waste products that suppress slow-growers [51].
- Media Toxicity: Standard agar media can contain hydrogen peroxide byproducts from autoclaving phosphate-agar mixtures, inhibiting growth of catalase-deficient species [51].
FAQ 2: How can I physically separate slow-growers from fast-growers?
Answer: Physical separation techniques are fundamental to reducing competition:
- Dilution-to-Extinction Culturing: Serial dilution of inocula to statistical endpoint in low-nutrient media reduces cell densities until individual cells grow in isolation, effectively separating taxa [1] [6].
- Double Emulsion Encapsulation: Single-cell encapsulation in water-in-oil-in-water double emulsions (GrowMiDE platform) creates nanoscale bioreactors that prevent direct competition through nutrient privatization [56].
- Diffusion Chambers: Semi-permeable chambers placed in natural environments allow nutrient exchange while physically separating species [1] [51].
- Membrane Filtration: Using pore-size-specific filters to separate cells based on physical dimensions [57].
FAQ 3: What media modifications support slow-growing microbes?
Answer: Media optimization is crucial for cultivating slow-growers:
- Nutrient Dilution: Using 10-100x diluted standard media or natural water/soil extracts reduces concentrations to oligotrophic levels [51].
- Extended Incubation: Incubation periods from weeks to months accommodate generation times of hours to days [1] [51].
- Supplementation: Adding catalase to neutralize Hâ‚‚Oâ‚‚ in agar media, soil extracts for growth factors, or resuscitation-promoting factors (Rpf) [51].
- Physiological Mimicry: Creating synthetic media that mirror natural environmental conditions (e.g., artificial urine medium, synthetic cystic fibrosis medium) [58].
FAQ 4: How can I verify I've isolated a novel slow-growing species?
Answer: Confirmation requires multiple approaches:
- 16S rRNA Sequencing: Phylogenetic identification against databases like SILVA or Greengenes [1] [6].
- Growth Kinetics: Quantifying specific growth rates in low-nutrient conditions confirms oligotrophic characteristics [56] [59].
- Microscopy: Cell morphology examination and viability staining [56].
- Metabolic Profiling: Substrate utilization patterns against phenotypic arrays [56].
Troubleshooting Common Experimental Issues
Problem: Continued Overgrowth by Fast-Growing Contaminants
Solutions:
- Increase dilution factor in extinction culturing
- Implement multiple rounds of double emulsion sorting [56]
- Add mild growth inhibitors specific to fast-growers (e.g., sodium pyruvate for some copiotrophs)
- Use flow cytometry with viability staining to detect slow-growers amidst contaminants [56]
Problem: No Growth Observed in Isolation Attempts
Solutions:
- Extend incubation time (weeks to months) with proper evaporation controls [51]
- Test multiple natural environment extracts as media supplements [51]
- Implement co-culture approaches with helper strains [1] [58]
- Verify media osmolality and pH match natural environment
Problem: Isolates Cannot Be Subcultured to Purity
Solutions:
- Maintain original environmental conditions during subculturing
- Use solid versus liquid media variations
- Add signaling molecules from original community (e.g., acyl-homoserine lactones)
- Attempt cultivation in simulated natural environment chambers [1]
Experimental Protocols
Principle: Encapsulates individual bacterial cells in picoliter-scale aqueous droplets surrounded by oil shell suspended in outer aqueous phase, physically separating organisms and their nutrient niches.
Materials:
- Microfluidic double emulsion generation device
- HFE7500 oil with 2.2% Ionic Krytox surfactant [56]
- Inner aqueous phase: Growth medium + 0.5% BSA
- Cell phase: Sample diluted to OD₆₀₀ = 0.05 + 10% Optiprep density modifier
- Outer aqueous phase: Growth medium + 2% Pluronic F68 + 1% Tween-20
Procedure:
- Prepare all phases anaerobically if required (3-day oxygen equilibration)
- Load phases into respective syringe pumps:
- Inner aqueous phase: 1000 μL/hr
- Cell phase: 1000 μL/hr
- Oil phase: 2000 μL/hr
- Outer aqueous phase: 6000 μL/hr
- Generate double emulsions into collection vial
- Incubate emulsions at appropriate temperature (weeks to months)
- Sort grown emulsions via FACS based on fluorescence or optical density
- Break emulsions and plate on solid media or sequence contents
Principle: Extreme dilution reduces cell density to point where most growth wells contain either zero or one cell, eliminating competition.
Materials:
- Sterile 96-well plates
- Low-nutrient media (10% R2A, 10% marine broth, or soil extract medium)
- Environmental sample (filter-concentrated if aquatic)
- Multichannel pipettes and sterile reservoirs
Procedure:
- Prepare dilution series of sample across 96-well plate (100μL/well)
- Incubate at in situ temperature for 3-12 weeks
- Monitor turbidity weekly visually or via plate reader
- From positive wells, subculture to fresh low-nutrient media
- Confirm purity via 16S rRNA sequencing and microscopy
Table 1: Performance Comparison of Isolation Techniques
| Technique | Recovery Rate | Incubation Time | Throughput | Key Advantages |
|---|---|---|---|---|
| Double Emulsion (GrowMiDE) [56] | 22x increase for Negativicutes | Weeks | High (10³ droplets/sec) | Nutrient privatization; FACS compatible |
| Dilution-to-Extinction [6] | ~14% of wells positive | 3-12 weeks | Medium (10³ cultures) | Simple equipment; high diversity recovery |
| Diffusion Chambers [1] | Up to 40% recovery | 2-8 weeks | Low (10¹ chambers) | In situ conditions; minimal lab manipulation |
| Co-culture Approaches [58] | Varies with partner | 1-4 weeks | Medium (10² co-cultures) | Provides growth factors; mimics nature |
Table 2: Media Formulations for Slow-Growing Microbes
| Media Type | Base Composition | Key Additives | Target Groups | Incubation |
|---|---|---|---|---|
| Diluted R2A [51] | 10% R2A broth | Soil extract (0.5%) | Soil oligotrophs | 4-8 weeks, 25°C |
| Artificial Seawater Medium [56] | Modified seawater salts | Lactate (60mM), sulfate (30mM) | Sulfate-reducers like D. ferrophilus | 2-4 weeks, 30°C |
| Synthetic CF Medium [58] | SCFM2 base | Amino acids, mucin (0.2g/L) | CF microbiome | 3-8 weeks, 37°C |
| Low-Nutrient Freshwater [6] | Filter-sterilized habitat water | Cycloheximide (50μg/mL) | Freshwater bacterioplankton | 4-12 weeks, in situ temp |
Research Reagent Solutions
Table 3: Essential Materials for Microbial Isolation
| Reagent/Material | Function | Application Examples |
|---|---|---|
| Ionic Krytox FSH Surfactant [56] | Stabilizes double emulsion interfaces | Prevents droplet coalescence in GrowMiDE |
| Pluronic F68 [56] | Stabilizes outer aqueous phase | Biocompatible surfactant for DE systems |
| Optiprep Density Modifier [56] | Equalizes phase densities during DE generation | Maintains stable flow rates in microfluidics |
| Resuscitation-Promoting Factor (Rpf) [6] | Reactivates dormant cells | Increases culturability from soil samples |
| Catalase Supplement [51] | Neutralizes Hâ‚‚Oâ‚‚ in agar media | Counteracts cultivation inhibition |
| Soil Extract Supplement | Provides unknown growth factors | Mimics natural nutrient conditions |
| Synthetic Cystic Fibrosis Medium (SCFM2) [58] | Replicates infection site conditions | Improves growth of clinical isolates |
Workflow Diagrams
Diagram 1: Double Emulsion Isolation Workflow
Diagram 2: Media Optimization Strategy
Advanced Applications in Drug Discovery
The isolation of previously unculturable microbes directly addresses the "discovery void" in antibiotic development, where no new antibiotic classes have been discovered since 1987 [1] [58]. Slow-growing organisms often possess unique secondary metabolite pathways honed through competition in nutrient-limited environments. The GrowMiDE platform has demonstrated particular success in cultivating Negativicutes and Methanobacteria from human gut samples - taxa with known metabolic capabilities but previously resistant to isolation [56].
Furthermore, understanding polymicrobial interactions through these isolation techniques reveals that antibiotic efficacy can be significantly altered in community contexts compared to pure cultures [58]. Some compounds with poor activity against monocultures show significantly increased potency in polymicrobial contexts, suggesting that cultivation of interacting slow-growing species may unlock novel antimicrobial strategies [58].
For researchers focused on unculturable microbial species, host-assisted cultivation represents a paradigm shift. This technique moves beyond traditional in vitro methods by leveraging the natural, complex environment provided by a living host plant to cultivate otherwise recalcitrant endophytic bacteria. These endophytes, residing within plant tissues without causing harm, are a vast reservoir of microbial diversity with significant potential for drug development and agricultural applications [60] [61]. Success in this field requires navigating specific experimental challenges, from initiating the symbiotic relationship to analyzing the results. This technical support center is designed to provide actionable troubleshooting guides and detailed protocols to help you reliably tap into this potential.
Troubleshooting Common Experimental Challenges
FAQ: Frequently Encountered Problems
Q1: Why am I getting low rates of endophytic colonization in my host plant? Low colonization rates often stem from improper inoculation techniques or incompatible host-microbe pairings. Ensure that the bacterial inoculum is applied to young, healthy root systems, preferably at the root apical zone where entry is easiest [61]. The use of a bacterial strain with known chemotaxis toward your host plant's root exudates will significantly improve colonization success.
Q2: My cultured endophytes are consistently overgrown by fast-growing contaminants. How can I mitigate this? Surface sterilization of plant tissues is critical. Implement a multi-step sterilization protocol using reagents like ethanol and sodium hypochlorite, with concentration and exposure time optimized for your specific plant tissue [61]. Furthermore, employing the microcapsule cultivation method can physically separate slow-growing target endophytes from fast-growing contaminants, giving them a competitive advantage [62].
Q3: How can I confirm that a recovered isolate is a true endophyte and not a surface contaminant? Confirmation requires rigorous validation. After surface sterilization, the internal tissue should be plated. A true endophyte will grow from the internal tissue, while a contaminant will not. You can further confirm endophytic colonization using molecular techniques like fluorescence in situ hybridization (FISH) with specific probes to visualize the bacteria within the plant tissue [61].
Q4: What are the main advantages of using semi-permeable devices like diffusion chambers or magnetic capsules? These devices allow you to cultivate microbes in situ by exposing them to the natural chemical and environmental signals of their native habitat (soil, seawater) while containing them for easy retrieval. Nutrients and growth factors diffuse in, but the cells are trapped, enabling the growth of microbes that require unknown or complex environmental conditions [1] [62].
Troubleshooting Guide Table
| Problem | Possible Cause | Recommended Solution |
|---|---|---|
| No growth from environmental samples | Lack of essential environmental signals or growth factors; competition. | Use in situ cultivation with diffusion chambers or magnetic capsules [1] [62]. |
| Low endophytic colonization efficiency | Incompatible host-microbe pair; incorrect inoculation method. | Screen for chemotaxis; inoculate at root apical zones or lateral root cracks [61]. |
| Overgrowth by contaminants | Inefficient surface sterilization of plant tissue. | Optimize sterilization protocol (e.g., ethanol concentration, exposure time); use microcapsules for separation [61] [62]. |
| Inability to subculture isolates | Dependence on specific host-derived nutrients or signals. | Use host-mimicking conditions; consider co-culture with a helper microbial strain [1]. |
| Unreliable plant growth promotion data | Uncontrolled environmental variables; inconsistent inoculum. | Standardize growth conditions and bacterial inoculum density; include appropriate control plants [60]. |
Essential Experimental Protocols
Protocol 1:In SituCultivation Using Magnetic Microcapsules
This protocol leverages recent technology to cultivate unculturable endophytes by encapsulating them in a semi-permeable, magnetic shell and returning them to their native environment [62].
- Cell Encapsulation: Mix a diluted suspension of cells (from surface-sterilized plant tissues or an environmental sample) with a polydimethylsiloxane (PDMS) solution containing magnetic iron oxide nanoparticles. Use a microfluidic device to form monodisperse, water-core microcapsules at a high throughput (e.g., ~6,000 per minute).
- Curing and Placement: Incubate the microcapsules at a mild temperature to cross-link the PDMS shell, forming a stable, semi-permeable membrane. Transfer the sealed microcapsules to a natural growth environment, such as sterile sand infused with a nutrient solution or directly into the soil adjacent to plant roots.
- Retrieval and Isolation: After an incubation period, recover the microcapsules from the medium using a magnet. To release the encapsulated microbes, transfer the capsules to pure water. The osmotic difference will cause the capsules to burst, freeing the cultured cells for plating and isolation.
Protocol 2: Establishing Endophyte-Host Symbiosis in a Gnotobiotic System
This protocol outlines the steps for axenically introducing a bacterial endophyte into a host plant to study the symbiotic interaction under controlled conditions.
- Plant Preparation: Surface-sterilize plant seeds using a sequence of 70% ethanol and a diluted sodium hypochlorite solution, followed by multiple rinses with sterile water. Germinate the seeds on sterile agar plates or in sterile vermiculite.
- Bacterial Inoculum Preparation: Grow the endophytic bacterial strain in a suitable liquid medium until the late logarithmic phase. Centrifuge the culture and resuspend the cells in a sterile buffer (e.g., 10 mM MgCl₂) to a standardized optical density (e.g., OD₆₀₀ = 0.5).
- Inoculation: For seedlings, carefully dip the intact roots into the bacterial suspension for a set duration (e.g., 30-60 minutes). Alternatively, apply the inoculum directly to the soil or growth medium near the roots of young plants.
- Confirmation of Colonization: After a suitable period (e.g., 1-2 weeks), harvest the plants. Surface-sterilize the roots/shoots again and homogenize the internal tissues in a sterile buffer. Plate the homogenate on selective media to re-isolate the bacterium and confirm successful colonization.
Signaling Pathways and Workflows
Endophyte Recognition and Nodulation
The following diagram illustrates the established signaling pathway in rhizobia-legume interactions, a classic model for host-assisted cultivation. This process shares conceptual parallels with how many beneficial endophytes initiate colonization.
Diagram Title: Endophyte Signaling and Nodulation
Host-Assisted Cultivation Workflow
This workflow integrates modern cultivation techniques with the study of endophyte function.
Diagram Title: Host-Assisted Cultivation Workflow
The Scientist's Toolkit: Research Reagent Solutions
Key Research Materials and Their Functions
| Research Reagent / Material | Function in Host-Assisted Cultivation |
|---|---|
| Magnetic PDMS Microcapsules [62] | Semi-permeable vessels for in situ cultivation; allow nutrient/waste exchange while physically protecting slow-growing endophytes and enabling easy magnetic retrieval. |
| Polydimethylsiloxane (PDMS) [62] | A polymer used to create the semi-permeable shell of microcapsules; its porosity is tunable by controlling cross-links during fabrication. |
| Diffusion Chambers [1] | Semi-permeable chambers placed in natural environments to allow chemical exchange, facilitating the growth of unculturable microbes by providing missing environmental factors. |
| 1-Aminocyclopropane-1-Carboxylate (ACC) [60] | A precursor to plant ethylene; used as a nitrogen source to select for and identify endophytes producing ACC deaminase, a key enzyme in plant stress tolerance. |
| ATCC Microbial Reference Strains [63] | Fully authenticated microbial materials crucial as positive controls in colonization experiments, assay development, and comparative genomic studies. |
| Siderophores (e.g., Pyochelin) [60] | Iron-chelating compounds produced by endophytes; their production is a key mechanism for biocontrol and can be a marker for beneficial plant-microbe interactions. |
| Plant Growth Media (e.g., N-free) | Selective media used to isolate and identify nitrogen-fixing endophytic bacteria, such as Azospirillum spp. and Azoarcus spp. [60]. |
Key Quantitative Data for Experimental Design
Efficacy of Advanced Cultivation Methods
The table below summarizes performance data for different cultivation techniques, providing benchmarks for your experimental planning.
| Cultivation Method | Reported Recovery Rate | Key Metric | Reference |
|---|---|---|---|
| Standard Petri Plates | ~0.05% | Cell recovery from marine sediment | [1] |
| Diffusion Chambers | Up to 40% | Cell recovery from marine sediment | [1] |
| Magnetic Microcapsules | ~50,000 cells from 5 cells | Culture density in nanoliter volume | [62] |
| Bacterial Endophytes (ACC deaminase) | N/A | Reduces abiotic stress by lowering plant ethylene levels | [60] |
| Phosphate-Solubilizing Endophytes | Up to 50% yield increase | Corn yield with reduced P fertilizer | [60] |
Common Endophyte Mechanisms and Impacts
This table outlines the primary mechanisms by which endophytes promote plant growth and their demonstrated effects.
| Mechanism of Action | Example Endophyte Genera | Documented Impact on Plants | |
|---|---|---|---|
| Nitrogen Fixation | Azospirillum, Azoarcus | Increased nitrogen availability for plant growth | [60] |
| Phosphate Solubilization | Achromobacter, Bacillus | Increased phosphorus availability, boosting yield | [60] |
| Phytohormone Production (IAA) | Pseudomonas | Direct stimulation of root and shoot growth | [60] [61] |
| Biocontrol (Antibiotic Production) | Burkholderia, Pseudomonas | Suppression of pathogenic microbes like Ralstonia solanacearum | [60] |
| Siderophore Production | Pseudomonas | Competition with pathogens for trace metals | [60] |
Troubleshooting Guides
FAQ: Addressing Common Cultivation Challenges
1. My bacterial cultures are not growing, despite using rich media. What could be wrong? The issue may be that the organisms have entered a difficult-to-cultivate state due to environmental stress. Research shows that a significant proportion of gut microbiota, once considered 'unculturable,' can be grown by recreating their specific environmental conditions [64]. Furthermore, anoxic (oxygen-free) conditioning can generate a subpopulation of bacteria, such as Pseudomonas aeruginosa and Staphylococcus aureus, that become difficult-to-culture (DTC) [65]. For organisms conditioned in low oxygen, try substituting molecular oxygen (Oâ‚‚) with an alternative electron acceptor like 10 mM nitrate (NO₃â») during plating. Adding ROS scavengers like sodium pyruvate or catalase to the growth medium can also resuscitate growth by countering oxidative damage encountered upon re-exposure to air [65].
2. How can I better control pH during procedures outside an incubator? Using a bicarbonate/CO₂ buffering system is ineffective in room atmosphere. For procedures like sample manipulation, ICSI, or cryopreservation, use a biological buffer in your handling media [66]. Zwitterionic buffers like HEPES (pKa 7.31 at 37°C) or MOPS (pKa 6.93 at 37°C) are commonly employed. Select a buffer with a pKa value close to your desired working pH for maximum buffering capacity. Be aware that the buffer itself can impact cellular processes, so empirical testing is recommended [66].
3. What are the signs of contamination in my bioreactor, and how can I find the source? Common signs include [32]:
- Unexpected growth kinetics: Growth occurs earlier than expected or has different characteristics.
- Visual changes: Increased turbidity or, for cell culture media with phenol red, a color change from pink to yellow (indicating acid formation).
- Unusual smells: The culture smell differs from previous, uncontaminated runs.
To troubleshoot [32]:
- Check the inoculum: Re-plate a sample of your seed train on a rich medium to check for hidden contaminants.
- Inspect sterilization procedures: Verify autoclave temperatures and ensure steam can penetrate all items. For persistent spore-forming contaminants, disassemble and repeatedly autoclave equipment.
- Examine components: Check all O-rings, seals, and sensors for damage and replace them regularly. Ensure exit gas filters are not wet, which can allow microbial grow-through.
4. Why is the incubation temperature critical for detecting certain contaminants? Some bacterial strains, like Pseudomonas fluorescens, have optimal growth at lower temperatures. A study on platelet concentrates found that while most common bacterial contaminants were detected faster at 35°C, one strain of P. fluorescens showed a faster time-to-detection at 25°C [67]. This suggests that for general screening, 35°C is sufficient, but if specific, cold-tolerant species are suspected, a lower incubation temperature should be considered.
Optimizing Key Environmental Parameters
Table 1: Fine-Tuning Temperature for Microbial Growth
| Temperature | Typical Use | Impact on Microbes | Application Example |
|---|---|---|---|
| High (Moist Heat) | Sterilization | Denatures proteins; cidal action. | Autoclaving (121°C, 15-45 min, 15 psi): Kills vegetative cells and endospores [68]. |
| High (Dry Heat) | Sterilization | Oxidizes proteins; cidal action. | Hot Air Oven (171°C for 1 hr): For sterilizing glassware and metal instruments [68]. |
| Ambient (20-25°C) | Cultivation | Supports growth of psychrophilic/mesophilic organisms. | Bacterial Screening: Incubation at 25°C can improve detection of some Pseudomonas species [67]. |
| Physiological (35-37°C) | Cultivation | Optimal for many human pathogens and mammalian cells. | Routine Diagnostics: Standard temperature for culturing many human-associated bacteria [67]. |
| Low (5°C to -10°C) | Storage | Slows or stops metabolic activity; static action. | Refrigeration/Freezing: Preserves cultures and food by inhibiting microbial growth [68]. |
Table 2: Selecting and Using Biological pH Buffers Source: [66]
| Buffer Name | pKa at 37°C | Temp. Effect (dpH/dT) | Ideal Working pH | Considerations for Use | |
|---|---|---|---|---|---|
| HEPES | 7.31 | -0.014 | 7.1 - 7.5 | Common choice for cell culture; widely tested. | |
| MOPS | 6.93 | -0.013 | 6.8 - 7.2 | Used in bacterial and yeast culture media. | |
| TES | 7.16 | -0.020 | 7.0 - 7.4 | ||
| PIPES | 6.66 | -0.008 | 6.5 - 7.0 |
Key Criteria for Buffer Selection: pKa near working pH, high water solubility, membrane impermeability, minimal salt effects, and chemical stability [66].
Table 3: Strategies for Managing Oxygen Requirements
| Condition | Description | Cultivation Strategy | Example |
|---|---|---|---|
| Strict Anaerobe | Killed or inhibited by Oâ‚‚. | Use anaerobic chambers, anaerobic jars, or specialized media with Oâ‚‚ scavengers. | Many gut Firmicutes and Bacteroidetes [64]. |
| Microaerophile | Requires low Oâ‚‚ (2-10%). | Use specialized gas-generating packs or controlled gas incubators. | Suggested state for some putative pathogens [69]. |
| Facultative Anaerobe | Grows with or without Oâ‚‚. | Can grow under aerobic or anaerobic conditions. | E. coli, Staphylococcus spp. [65]. |
| Difficult-to-Culture (DTC) | Loss of culturability after stress. | Provide alternative electron acceptors (e.g., 10 mM NO₃â») or ROS scavengers (catalase, pyruvate). | P. aeruginosa after anoxic conditioning [65]. |
Experimental Protocol: Resuscitating Difficult-to-Culture (DTC) Bacteria from Anoxic Conditioning
This protocol is adapted from research demonstrating that anoxic conditioning can render bacteria like P. aeruginosa and S. aureus difficult-to-culture, and outlines methods for their resuscitation [65].
1. Materials
- Anaerobic chamber (with Nâ‚‚/Hâ‚‚/COâ‚‚ atmosphere)
- Standard and nitrate-supplemented (10 mM KNO₃) LB agar plates
- Catalase solution
- Sodium pyruvate
- 0.9% NaCl for dilutions
2. Method
- Anoxic Conditioning: Grow the bacterial strain (e.g., P. aeruginosa PAO1) in a biofilm or planktonic culture under anoxic conditions in the anaerobic chamber for a set period.
- Sample Preparation: Harvest the cells and sonicate if necessary to break up aggregates. Perform a ten-fold dilution series in 0.9% NaCl inside the anaerobic chamber.
- Viability Plating:
- Anoxic Plating: Plate diluted samples onto LB agar plates supplemented with 10 mM NO₃â». Incubate plates inside the anaerobic chamber at 37°C for 2 days.
- Normoxic Plating: Using the same dilution series, plate samples on LB plates (with or without 10 mM NO₃â»). Incubate these plates aerobically (normoxically) at 37°C for 1 day, followed by 1 day at room temperature.
- Resuscitation Plating: To test for DTC cells due to ROS sensitivity, add the ROS scavenger catalase (or 0.1% sodium pyruvate) to the normoxic LB plates before plating the anoxically-conditioned samples. Incubate aerobically.
- Analysis: Count Colony Forming Units (CFU/mL) from all conditions. The difference in counts between anoxic plating and normoxic plating indicates the DTC subpopulation. Successful growth on plates with catalase or pyruvate confirms oxidative stress as a key factor.
Visualizing the Workflow and Key Concepts
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Reagents for Cultivating Fastidious and DTC Organisms
| Reagent | Function/Explanation | Example Application |
|---|---|---|
| YCFA Medium | A complex, broad-range bacteriological medium. | Supported the cultivation of 137 distinct bacterial species from human gut microbiota, including 45 novel species [64]. |
| Nitrate (NO₃â») | Serves as an alternative terminal electron acceptor for anaerobic respiration. | Resuscitated DTC P. aeruginosa and staphylococci when used in plating agar after anoxic conditioning [65]. |
| Catalase | An enzyme that decomposes hydrogen peroxide (Hâ‚‚Oâ‚‚), a reactive oxygen species (ROS). | Added to growth media to counteract oxidative stress, improving the culturability of DTC organisms upon re-exposure to air [65]. |
| Sodium Pyruvate | A ROS scavenger that helps neutralize hydrogen peroxide. | Functions similarly to catalase; inclusion in culture media can enhance the recovery of stressed cells [65]. |
| Biological Buffers (HEPES/MOPS) | Zwitterionic buffers that maintain stable pH outside a COâ‚‚ incubator. | Used in handling media for procedures like gamete/embryo manipulation, ICSI, and cryopreservation to minimize pH fluctuations [66]. |
| Ethanol | Used as a selective agent for bacterial spores. | Ethanol treatment (e.g., 70%) can select for ethanol-resistant spores from a mixed population, enriching for spore-forming bacteria [64]. |
| Taurocholate | A bile salt that acts as a potent germinant for bacterial spores. | Triggers germination of commensal spore-forming bacteria, increasing culturability by 8 to 70,000-fold [64]. |
Why is microbial cultivation important in the age of metagenomics? While metagenomic sequencing has vastly expanded our understanding of microbial diversity, the majority of environmental microbes remain uncultured. Cultivation is crucial for directly studying and confirming the metabolic and physiological functions, and ecological roles of these microbes [13]. Pure cultures allow for experimental validation of hypotheses generated from genomic data and can find applications in new probiotics, biocontrol agents, and industrial processes [13].
What is the role of multi-omics in guiding cultivation? Multi-omic studies, which integrate data from various layers like the genome, transcriptome, epigenome, metabolome, and proteome, provide a deeper, systems-level insight into host-microbe and microbe-microbe interactions [70]. The growing abundance of metagenomic and meta-transcriptomic sequence information from various environments provides more opportunities to guide the targeted isolation and cultivation of microbes of interest by revealing their predicted metabolic requirements and dependencies [13].
What are the common analytical approaches for multi-omics data? A common analysis involves identifying differentially abundant features (e.g., species, metabolites) between sample groups. More advanced intermediate integration methods, like the "MintTea" framework, are being developed to identify disease-associated multi-omic modules. These modules comprise features from multiple omics (e.g., specific bacterial species and correlated metabolites) that shift in concert and collectively associate with a condition, providing a more coherent hypothesis for underlying mechanisms [71].
Troubleshooting Guides
Pre-Cultivation Planning
Problem: How to select target microbes from complex metagenomic data?
- Challenge: Metagenomic data can contain thousands of microbial species. Selecting which rare or uncharacterized groups to target for cultivation is a key first step.
- Solution:
- Generate Metagenome-Assembled Genomes (MAGs): Perform high-quality metagenomic assembly and binning to reconstruct genomes from uncultured organisms [13]. This provides a genomic basis for understanding the target's potential.
- Prioritize Unexplored Diversity: Focus on phylogenetic lineages that have no cultivated representatives or are consistently detected but poorly characterized in your environment of interest [13] [72].
- Identify Functional Potential: Analyze MAGs for unique metabolic pathways, such as the ability to degrade specific compounds or perform novel syntrophic interactions, that make them ecologically significant [13].
Problem: How to design culture media based on genomic and metabolomic data?
- Challenge: The standard, nutrient-rich media used in labs often do not support the growth of many environmental microbes, which may have specialized or minimal nutrient requirements [13].
- Solution:
- Infer Metabolism from MAGs: Use genomic data to predict the carbon, nitrogen, and energy sources a microbe can utilize. Look for complete pathways and transporter genes [13].
- Leverage Co-occurrence Data: If a target microbe consistently co-occurs with other species in metagenomic profiles, it may depend on cross-feeding. Consider including predicted metabolic byproducts from the partner in your media [13].
- Use Metabolomics to Define the Environment: Analyze the metabolomic profile of the natural sample. The metabolites present (e.g., sugars, amino acids, organic acids) provide direct clues about the nutrients available and consumed in the native habitat [70] [71]. Design media that mimic this chemical environment.
Cultivation Execution & Optimization
Problem: How to address slow growth and low abundance?
- Challenge: Target microbes may be slow-growing and get outcompeted by fast-growing species in a standard cultivation setup, or they may exist in very low abundances in the inoculum [13].
- Solution:
- Use Dilution-to-Extinction: Diluting the inoculum in a very low-nutrient medium can reduce competition from fast-growers and allow rare microbes to proliferate [13].
- Apply High-Throughput Culturomics: Use robotic systems to test hundreds of different media conditions and micro-environments in parallel to increase the chances of capturing the target [13].
- Incorporate Specific Substrates: Based on genomic predictions, supplement media with specific sugars, polymers, or other compounds that only the target microbe is predicted to use, thereby giving it a selective advantage [13].
Problem: How to cultivate microbes dependent on metabolic interactions?
- Challenge: Some microbes require specific growth factors or metabolic byproducts from other community members and will not grow in isolation [13].
- Solution:
- Start with Enrichment or Co-Cultures: Initially, aim for enrichment cultures that increase the population of a microbial group. Stable defined co-cultures (e.g., the target with a single helper strain) are also valuable for gaining biological insights and maintaining the organism [13].
- Use Metabolomics to Identify Dependencies: Analyze spent media from successful co-cultures with metabolomics to identify the metabolites that are being consumed and produced, which can reveal the nature of the cross-feeding [70].
- Feed Predicted Cross-Feeding Metabolites: If genomic analysis suggests the target microbe lacks a pathway for synthesizing an essential vitamin or amino acid, provide that compound directly in the medium [13].
Key Reagents & Materials
The table below lists essential materials and their functions for a multi-omics-guided cultivation pipeline.
Table 1: Research Reagent Solutions for Multi-Omics-Guided Cultivation
| Item | Function in Experiment |
|---|---|
| DNA Extraction Kit (for complex samples) | To extract high-quality, high-molecular-weight DNA from environmental samples for shotgun metagenomic sequencing. |
| Metagenomic Sequencing Reagents | For generating the raw DNA sequence data used for taxonomic profiling and Metagenome-Assembled Genome (MAG) reconstruction. |
| Mass Spectrometry-Grade Solvents | For metabolomic sample preparation and analysis using Liquid Chromatography-Mass Spectrometry (LC-MS). |
| Custom Culture Media Components | To formulate tailored media based on genomic and metabolomic inferences, including specific carbon/nitrogen sources, vitamins, and minerals. |
| Anaerobic Chamber/Gas Pack Systems | To cultivate microbes originating from anaerobic environments (e.g., gut, sediments) by maintaining an oxygen-free atmosphere. |
| High-Throughput Cultivation Equipment | Such as automated liquid handlers and microplate readers, to test hundreds of media conditions in parallel (culturomics). |
| Reference Genomic Databases | (e.g., KEGG, GenBank) For annotating functional genes and metabolic pathways in reconstructed MAGs [13]. |
| Metabolomics Reference Libraries | (e.g., HMDB, METLIN) For identifying metabolites detected in the natural sample or in spent culture media [70]. |
Experimental Protocols & Data Integration
Core Workflow for Multi-Omics-Informed Cultivation
The following diagram illustrates the integrated workflow from sample collection to pure culture, highlighting how data from different omics layers informs each step.
Protocol: Targeted Isolation Using Genomically-Inferred Substrates
This protocol provides a detailed methodology for using genomic predictions to cultivate a target microbe.
- Objective: To isolate a specific, uncultivated microbial target by providing it with a predicted essential substrate as a selective advantage.
- Principles: This method leverages the analysis of Metagenome-Assembled Genomes (MAGs) to identify metabolic capabilities that are unique to the target organism. By formulating a culture medium that contains a substrate that only the target can utilize, you can selectively enrich for its growth [13].
- Procedure:
- Step 1: Genomic Prediction
- Reconstruct a MAG for your target microbe from metagenomic data.
- Annotate the genome to identify complete metabolic pathways. Pay special attention to pathways for complex carbohydrate degradation, unique electron acceptors, or specific vitamin synthesis.
- Key Decision Point: Select a substrate that the target possesses the genes to utilize, but that is expected to be unusable by the majority of other, fast-growing microbes in the inoculum.
- Step 2: Media Formulation
- Prepare a basal mineral medium that mimics the ionic strength and pH of the source environment.
- Supplement this basal medium with the predicted substrate as the sole or primary carbon and energy source.
- Troubleshooting Tip: Test multiple concentrations of the substrate (e.g., 0.01%, 0.1%, 1.0%) and parallel media with different predicted substrates, as the genomic prediction may be imperfect.
- Step 3: Inoculation and Incubation
- Inoculate the media with a diluted sample of the original environmental community. Dilution helps reduce competition.
- Incurate under conditions (temperature, atmosphere) that match the source habitat.
- Monitor growth over an extended period (weeks to months), as target organisms may be slow-growing [13].
- Step 4: Detection and Isolation
- Use target-specific PCR primers (designed from the MAG) to screen for the presence of the microbe in enrichment cultures showing growth.
- Once a positive enrichment is identified, proceed to pure culture isolation using standard techniques like streak-plating on solid media formulated with the same selective substrate.
- Step 1: Genomic Prediction
Data Interpretation & Validation FAQ
How do I know if my cultivation strategy is working? Success is measured by a sustained increase in the relative abundance of the target microbe in enrichment cultures, which can be tracked using qPCR with primers specific to the target (designed from its MAG) or by metagenomic sequencing of the enrichment community. Ultimately, obtaining a stable pure culture is the definitive proof of success [13].
The microbe grew in co-culture but not alone. What next? This is a common and valuable outcome. To progress:
- Identify the Helper Strain: Sequence the genome of the helper strain to understand its metabolism.
- Analyze the Interaction: Use metabolomics to compare the spent media from the co-culture versus pure cultures of the helper strain. This can reveal the essential metabolite(s) or growth factor(s) being provided [70] [71].
- Refine the Medium: Attempt to replace the helper strain by supplementing the culture medium with the identified essential metabolite.
How can I validate the functional role of a cultivated novel microbe? Once a pure culture is obtained, you can:
- Sequence its Genome: Compare the isolate's genome to the original MAG to confirm they match.
- Conduct Phenotypic Assays: Test its ability to utilize the substrates that were predicted from its genome.
- Re-introduce to a System: In a model system (e.g., gnotobiotic mouse, simplified microbial community), study the effect of the introduced isolate on community structure and function, thereby confirming its hypothesized ecological role [13].
Validating Cultivation Success: From Genomic Fidelity to Functional Discovery
Frequently Asked Questions (FAQs)
FAQ 1: Why is it important to compare cultured strains to Metagenome-Assembled Genomes (MAGs)?
While MAGs have dramatically expanded our map of microbial diversity, they are not a perfect representation of an organism's complete genetic blueprint. Comparisons with cultured isolates are crucial for validation. A 2025 study on Klebsiella pneumoniae found that over 60% of gut-associated MAGs belonged to new sequence types missing from culture collections, and integrating MAGs nearly doubled the perceived phylogenetic diversity of this species [73]. Cultured strains enable researchers to close genomic gaps, confirm the existence and structure of predicted metabolic pathways, and directly link genotype to phenotype through experiments that are impossible with MAGs alone [6] [10].
FAQ 2: What are the common genomic discrepancies between a cultured isolate and its closely related MAG?
Discrepancies often arise from the fundamental differences in how these genomes are reconstructed. Common issues include:
- Fragmentation and Incompleteness: MAGs are often fragmented into many contigs, potentially missing or misassembling regions like plasmids, repeat sequences, or genomic islands [6].
- Gene Misannotation: MAGs may contain genes annotated as "hypothetical proteins." If these genes have an associated EC number, it is a common validation error that must be corrected by either removing the EC number or providing a valid product name [74].
- Contamination and Chimerism: MAGs can be contaminated with foreign DNA from other organisms in the metagenomic sample, leading to a chimeric genome that does not represent a single organism [73].
- Strain Heterogeneity: A MAG may represent a composite of multiple, closely related strains from the same sample, whereas a cultured isolate represents a single strain [73].
FAQ 3: My MAG suggests the organism has specific metabolic capabilities (e.g., methylotrophy), but my cultured strain won't grow in the corresponding conditions. What should I check?
This is a common challenge in bridging genomic predictions with laboratory cultivation. Your troubleshooting should focus on:
- Growth Medium Design: Ensure your cultivation medium accurately replicates the natural environment. The use of defined, low-nutrient media that mimic environmental conditions (e.g., low carbon concentrations in µM range) has been key to cultivating previously uncultured oligotrophs [10].
- Essential Growth Factors: The organism might depend on co-occurring microbes for essential nutrients or detoxification of metabolites [10]. Consider using culture supernatants from helper strains or adding specific growth factors like the resuscitation-promoting factor (Rpf) to stimulate growth [1] [6].
- Genetic Code and Gene Start Sites: During genome annotation, verify that the correct genetic code was used. For prokaryotes, this is typically genetic code 11. An incorrect code can lead to false predictions of internal stop codons and mis-annotated genes [74].
Troubleshooting Common Validation Errors
When comparing the genome of your cultured strain to a MAG, you may encounter specific bioinformatic and annotation challenges. The table below summarizes common issues and their solutions.
Table 1: Troubleshooting Guide for Genomic Validation and Submission
| Error or Issue | Possible Cause | Solution and Fix |
|---|---|---|
| Internal Stop Codon in CDS [74] | Incorrect genetic code; mis-annotated start site; genuine pseudogene. | Force the use of the prokaryotic genetic code (gcode=11); adjust the codon_start qualifier; if the sequence is truly non-functional, add the /pseudo qualifier. |
| "Hypothetical protein" with EC number [74] | An Enzyme Commission number was included for a protein with an unknown or generic name. | Remove the EC number if the protein is truly uncharacterized. If the EC number is correct, use it to find and assign a valid, specific product name. |
| Poor Genome Assembly Quality | High fragmentation; many runs of ambiguous bases (N's). | Do not simply remove N's. Instead, label runs of 100 or more N's as assembly_gap with appropriate linkage evidence [74]. |
| Invalid Geographic Location [74] | The geographic location name (geo_loc_name) is not on the approved list or is formatted incorrectly. |
Use an approved geographic location name (e.g., "USA: Massachusetts"). More specific info can be added after a colon. Format: <approved name>: <specific area>. |
| Culture Collection Format Error [74] | The culture collection identifier is missing, has an unregistered institute code, or is incorrectly structured. | Format must be: <institution-code>:<culture id> (e.g., DSMZ:1234). Ensure the institution code is on the approved list. |
Experimental Protocols for Targeted Cultivation
Leveraging information from MAGs to guide the cultivation of uncultured species is a powerful strategy. Below are two key methodologies.
Protocol 1: High-Throughput Dilution-to-Extinction Cultivation
This method is highly effective for isolating slow-growing, oligotrophic bacteria that are outcompeted in standard rich media [10].
- Medium Preparation: Prepare a defined, low-nutrient medium that mimics the natural environment of the target organism (e.g., freshwater lakes). Carbon sources should be in the µM range. Autoclaved environmental water can be used, but defined media ensure reproducibility [10].
- Sample Inoculation: Serially dilute the environmental sample (e.g., water, soil suspension) to a theoretical concentration of approximately one cell per well in 96-deep-well plates.
- Incubation: Incubate the plates undisturbed for 6-8 weeks or longer at a temperature matching the source environment. Avoid agitation to prevent disturbing slow-growing microcolonies.
- Growth Screening: Screen wells for turbidity or use flow cytometry to detect growth. The goal is to obtain axenic cultures directly without enrichment.
- Strain Validation: Transfer positive cultures to fresh medium. Check for purity via 16S rRNA gene sequencing and microscopy. A 2025 study on freshwater microbes using this approach achieved a 12.6% viability rate, isolating 627 axenic strains from 14 lakes [10].
Protocol 2: Using Signaling Molecules for Resuscitation
Some bacteria require specific chemical signals to exit a dormant state and resume growth [1] [6].
- Prepare Supernatant: Grow a "helper" strain like Micrococcus luteus (which produces the resuscitation-promoting factor Rpf) in a suitable liquid medium.
- Harvest and Filter: Centrifuge the culture to pellet the cells and pass the supernatant through a 0.22 µm filter to remove all remaining bacteria.
- Supplement Media: Add a small volume (e.g., 1-10%) of the filter-sterilized supernatant to your basal cultivation medium.
- Inoculate and Incubate: Inoculate the supplemented medium with the environmental sample and incubate. Compare the diversity and number of colonies with a control plate without the supernatant. One study found that this treatment allowed the cultivation of 51 previously uncultured bacterial species from soil [6].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Cultivation and Genomic Validation
| Item | Function/Benefit |
|---|---|
| Defined Oligotrophic Media [10] | Mimics natural nutrient-scarce conditions, favoring the growth of previously uncultured oligotrophic microbes over fast-growing copiotrophs. |
| Resuscitation-Promoting Factor (Rpf) [6] | A bacterial cytokine that stimulates the resuscitation of dormant cells, increasing culturalbility from complex environments. |
| Diffusion Chambers / iChips [1] | A device that allows environmental nutrients and factors to diffuse into a chamber containing cells, effectively "culturing in situ." |
| NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [74] | A standardized, automated tool for annotating prokaryotic genomes upon submission to GenBank, ensuring consistency and reducing annotation errors. |
Workflow Diagram for MAG and Culture Comparison
The following diagram illustrates the integrated workflow for comparing cultured strains to MAGs, from sample to validated genome.
Cultivation Strategies for the "Uncultured Majority"
The diagram below outlines the decision process for selecting appropriate cultivation techniques based on genomic and ecological clues.
Long-read sequencing technologies from PacBio and Oxford Nanopore Technologies (ONT) are revolutionizing microbial genomics by generating reads thousands to tens of thousands of bases long from single DNA molecules. [75] This technological advancement is particularly transformative for studying unculturable microbial species, which may constitute the majority of microbial diversity. Unlike short-read technologies that struggle with repetitive regions and complex genomic architectures, long reads provide the continuity and context needed to assemble complete, high-quality microbial genomes directly from environmental samples. [76] [77] For researchers focused on improving microbial cultivation techniques, long-read sequencing of metagenomes offers a parallel path to genomic discovery, revealing the genetic potential of microbes that have thus far resisted laboratory cultivation.
Key Advantages for Unculturable Species Research
- Resolution of Repetitive Elements: Long reads can span repetitive genomic regions such as ribosomal RNA operons, transposons, and CRISPR-Cas arrays, which are often fragmented in short-read assemblies. This leads to more complete genomes and enables the study of important functional elements. [78]
- Access to Complex Genomic Regions: Technologies like PacBio HiFi sequencing provide uniform coverage, allowing researchers to analyze hard-to-sequence regions with high AT/GC content, long homopolymers, and palindromic sequences that are often inaccessible with other technologies. [77] [79]
- Epigenetic Profiling: Sequencing native DNA without amplification allows for the simultaneous detection of base modifications (e.g., methylation) alongside the primary genetic sequence. This provides insights into regulatory mechanisms in uncultured organisms. [80]
- Improved Binning and Classification: Longer contigs facilitate more accurate binning of metagenome-assembled genomes (MAGs) and better taxonomic classification, expanding the known microbial tree of life. [78] [81]
Experimental Protocols for Genome-Resolved Metagenomics
The following section details specific methodologies that have successfully generated high-quality microbial genomes from complex samples.
Protocol 1: Deep Long-Read Sequencing for Terrestrial Microbial Diversity
This protocol, derived from the Microflora Danica project, is designed for large-scale genome recovery from highly complex environments like soil and sediment. [78] [81]
- Sample Input: 154 complex environmental samples (125 soil, 28 sediment, 1 water).
- Sequencing Technology: Deep long-read Nanopore sequencing.
- Sequencing Depth: ~100 Gbp per sample (median), totaling 14.4 Tbp of data.
- Key Bioinformatic Workflow:
mmlong2, a custom workflow featuring:- Multi-coverage binning: Incorporates read mapping information from multiple samples.
- Ensemble binning: Applies multiple binning tools to the same metagenome.
- Iterative binning: Repeatedly bins the metagenome to recover additional MAGs.
- Outcome: The workflow recovered 23,843 MAGs, which were dereplicated into 15,640 species-level MAGs. Of these, 15,314 represented previously undescribed species, spanning 1,086 novel genera and expanding the phylogenetic diversity of prokaryotes by 8%. [78] [81]
Protocol 2: Hybrid Sequencing for Closing Genomes of Low-Abundance Organisms
This protocol, used to close the genome of an unculturable cable bacterium, combines long- and short-read sequencing to achieve high accuracy and completeness, ideal for low-abundance keystone organisms. [82]
- Sample Preparation: Generation of a clonal enrichment culture from a single filament to reduce strain diversity.
- DNA Extraction: High molecular weight DNA extraction, critical for long-read sequencing.
- Sequencing Technology: Hybrid approach using both Nanopore long-read and Illumina short-read shotgun sequencing.
- Assembly: Combined assembly of long and short reads to produce a circular, complete metagenome-assembled genome (MAG).
- Outcome: Recovery of a 5.09 Mbp circular MAG of a novel cable bacterium species, Candidatus Electrothrix scaldis, containing 1,109 previously unidentified genes. [82]
Protocol 3: Pangaea Workflow for Assembling Short-Reads with Long-Range Connectivity
For projects where cost-effective short-reads are preferred, the Pangaea assembler leverages long-range connectivity from linked-reads or by aligning short-reads to long-reads. [83]
- Input Data: Short-reads with physical barcodes (e.g., linked-reads) or virtual barcodes generated by alignment to long-reads.
- Core Workflow:
- Co-barcoded read binning: Uses a deep learning algorithm to group reads from the same genomic region.
- Multi-thresholding reassembly: Refines assembly of low-abundance microbes by gradually removing high-abundance data.
- Ensemble assembly: Integrates assemblies from different modules to improve contiguity.
- Outcome: Achieves higher contig continuity and recovers more near-complete MAGs (NCMAGs) than standard short-read assemblers, including some complete, circular genomes from human gut microbiomes. [83]
Research Reagent Solutions
The table below lists essential reagents and kits used in successful long-read metagenomic studies for unculturable microbial research.
| Item | Function | Example Products / Protocols |
|---|---|---|
| High Molecular Weight (HMW) DNA Extraction Kit | To obtain pure, long-strand DNA without shearing, which is crucial for long-read sequencing. | Circulomics Nanobind Big DNA Kit, QIAGEN Genomic-tip, QIAGEN Gentra Puregene Kit, QIAGEN MagAttract HMW DNA Kit [75] |
| Library Preparation Kit | To prepare DNA fragments for sequencing by repairing ends, adding adapters, and (in some cases) amplifying. | ONT Ligation Sequencing Kit, ONT Rapid Library Prep, PacBio SMRTbell Prep Kit [75] [79] |
| Low-Input Protocol | To enable sequencing from minimal sample input, such as low-biomass environmental samples. | PacBio Ampli-Fi protocol (for inputs as low as 1 ng) [79] |
| Barcoding/Multiplexing Kits | To pool multiple samples in a single sequencing run, reducing cost per sample. | PacBio Multiplexing Kits, ONT Barcoding Kits [79] [80] |
| Bioinformatic Tools | For basecalling, assembly, binning, and quality control of long-read data. | mmlong2 [78], Pangaea [83], SMRT Link (PacBio) [80], Guppy (ONT) [76], hifiasm-meta, metaFlye [79] |
Frequently Asked Questions (FAQs)
Q1: What are the main differences between PacBio and Oxford Nanopore long-read technologies?
Both technologies generate long reads but use fundamentally different principles. PacBio HiFi sequencing uses a circular consensus sequencing (CCS) approach, where a DNA molecule is sequenced multiple times to produce highly accurate reads (99.9%). [77] [80] Oxford Nanopore Technologies (ONT) measures changes in electrical current as DNA strands pass through a nanopore. ONT can produce ultra-long reads (over 100 kb) and is highly scalable from portable (MinION) to benchtop (PromethION) scales. [75] [84] The choice depends on the project's needs: HiFi is often preferred for its high accuracy, while ONT offers flexibility and the longest read lengths.
Q2: My microbial sample has very low biomass. Can I still use long-read sequencing?
Yes. Recent protocol developments have made this possible. For instance, the PacBio Ampli-Fi protocol supports library preparation from as little as 1 nanogram of DNA. [79] This is particularly useful for challenging samples with low biomass, degraded DNA, or contaminants. While amplification may be necessary, it still allows for the generation of high-quality metagenome-assembled genomes that would be difficult to obtain otherwise.
Q3: Why is my long-read assembly still fragmented, and how can I improve it?
Fragmentation can occur due to several factors:
- Insufficient Sequencing Depth: Highly complex communities or low-abundance species may require deeper sequencing. The Microflora Danica project sequenced to ~100 Gbp per sample. [78]
- High Microdiversity: The presence of many closely related strains can confuse assemblers. [78] Using a clonal enrichment strategy, as done for cable bacteria, can mitigate this. [82]
- Suboptimal DNA Quality: If the input DNA is sheared or degraded, read lengths will be shorter. Always use extraction methods that preserve high molecular weight DNA. [75]
- Bioinformatic Limitations: Try advanced assemblers or workflows like
Pangaea[83] ormmlong2[78] that are specifically designed for complex metagenomes.
Q4: How does long-read sequencing directly benefit the study of unculturable microbes?
Long-read sequencing bypasses the need for cultivation by allowing researchers to:
- Recover complete or near-complete genomes directly from environmental samples (MAGs). [78] [82]
- Identify novel genes and metabolic pathways that reveal the physiological potential of uncultured organisms. [82]
- Study epigenetic markers like methylation, which can provide clues about gene regulation. [80]
- Accurately reconstruct ribosomal RNA operons and other repetitive genetic elements that are key for phylogenetic classification. [78] This genomic information is invaluable for designing targeted cultivation strategies based on an organism's predicted metabolic needs.
Troubleshooting Guides
Issue: Low Yield of High-Quality Metagenome-Assembled Genomes (MAGs)
| Symptom | Possible Cause | Solution |
|---|---|---|
| Few HQ/MQ MAGs recovered after binning. | Insufficient sequencing depth for the sample's complexity. | Sequence deeper. For highly complex soils, aim for 100 Gbp or more per sample. [78] |
| High microbial diversity and evenness, with no dominant species. | Use iterative and multi-sample (co-assembly) binning techniques like those in the mmlong2 workflow to leverage coverage information across samples. [78] |
|
| High microdiversity within species (many similar strains). | Consider methods to reduce complexity, such as physical cell sorting or the establishment of clonal enrichment cultures prior to DNA extraction. [82] |
Issue: Poor DNA Quality and Quantity
| Symptom | Possible Cause | Solution |
|---|---|---|
| DNA is sheared (< 50 kb), low yield. | Harsh or inappropriate DNA extraction method. | Switch to a gentle, HMW-DNA specific kit (e.g., Circulomics Nanobind, QIAGEN Genomic-tip). [75] |
| Excessive pipetting or vortexing during library prep. | Pipette slowly and use wide-bore tips to minimize shearing forces. [75] | |
| Multiple freeze-thaw cycles or exposure to degrading agents. | Aliquot DNA and avoid freeze-thaw cycles. Ensure no denaturants, detergents, or chelating agents are present. [75] |
Issue: Short Read Lengths and Low Assembly Continuity
| Symptom | Possible Cause | Solution |
|---|---|---|
| Low read N50; assembly has low contig N50. | DNA is fragmented before or during extraction. | Optimize sample preservation and DNA extraction protocol for your specific sample type (e.g., soil, sediment). |
| Suboptimal sequencing chemistry or run conditions. | Follow manufacturer guidelines for library preparation and use the latest flow cells (e.g., ONT R10.4.1, PacBio Revio SMRT Cells). [75] [80] | |
| Complex metagenome with many repetitive regions. | Employ a hybrid assembly approach combining long reads with accurate short reads to polish the assembly, or use a HiFi sequencing approach for inherent accuracy. [82] [77] |
Workflow Visualization: Long-Read Metagenomics for Unculturable Species
The diagram below illustrates a generalized, end-to-end workflow for recovering high-quality microbial genomes from complex environmental samples using long-read sequencing.
Figure 1. End-to-end workflow for genome-resolved metagenomics. This pipeline outlines the key stages from sample collection to biological discovery, highlighting how long-read sequencing integrates into the study of unculturable microorganisms. HMW: High Molecular Weight; MAG: Metagenome-Assembled Genome; QC: Quality Control.
The discovery of novel antibiotics had largely stalled for decades, primarily because an estimated 99% of all bacterial species could not be cultured in traditional laboratory settings [85] [86]. This "microbial dark matter" represented a vast reservoir of potential therapeutic compounds that was technically inaccessible to researchers [87] [85]. The breakthrough leading to the isolation of Clovibactin came from addressing this fundamental cultivation problem, paving the way for accessing a new generation of antibiotics with novel mechanisms of action [88] [86].
Technical Solutions & Methodologies
Key Research Reagent Solutions
The following table details essential materials and reagents that formed the core toolkit for the discovery and characterization of Clovibactin.
Table 1: Essential Research Reagents and Tools for Unculturable Bacterium Research
| Tool/Reagent | Function in Research | Key Application in Clovibactin Discovery |
|---|---|---|
| iChip (Isolation Chip) | Enables cultivation of unculturable bacteria in their natural environment [89] [85]. | Cultivation of Eleftheria terrae ssp. carolina from sandy soil [85] [86]. |
| Solid-State NMR | Elucidates structure and mechanism of action of membrane-bound antibiotics under native-like conditions [90] [91]. | Determined Clovibactin's "cage-like" binding to pyrophosphate moiety [91] [86]. |
| Atomic Force Microscopy (AFM) | Visualizes nanostructures formed on bacterial membrane surfaces [90] [91]. | Revealed formation of stable supramolecular fibrils on bacterial membranes [90] [91]. |
| Bioassay-Guided Fractionation (HPLC) | Separates complex extracts and identifies active compounds based on biological activity [88]. | Isolated active Clovibactin fraction from fermented broth [92] [88]. |
| Mass Spectrometry | Determines precise molecular mass and aids in structural resolution [92] [88]. | Identified Clovibactin's unique mass of 903.5291 [M+H]+ [88]. |
Experimental Workflow: From Soil to Mechanism
The following diagram maps the core experimental workflow, from cultivating the unculturable bacterium to deciphering Clovibactin's unique mechanism of action.
Troubleshooting Guides & FAQs
This section addresses specific technical challenges and procedural questions that researchers may encounter when working with unculturable bacteria or characterizing novel antibiotics.
Cultivation and Isolation
Q1: Our iChip wells show no microbial growth even after several weeks. What could be the issue?
- A: Unculturable bacteria often have slow, finicky growth cycles.
- Check Incubation Time: Some species, like the producer of Clovibactin, were detected only after 12 weeks (approx. 3 months) of incubation [88]. Extend your observation period and monitor wells regularly under a dissecting microscope.
- Verify Nutrient Diffusion: Ensure the iChip's diffusion membranes are not clogged and the device is returned to an environment similar to the sample's origin (e.g., soil, water) to allow natural nutrients and growth factors to diffuse [89].
- Sample Pre-treatment: For accessing diverse actinomycetes, consider a mild heat pretreatment (e.g., 65°C for 30 minutes) to select for spore-forming bacteria [88].
Q2: We have a mixed culture producer. How can we simplify the isolation of a novel compound?
- A: If genome sequencing reveals a known antibiotic gene cluster in your producer, consider genetic disruption to shunt metabolic flux.
- Procedure: Identify the key biosynthetic gene cluster (BGC) via whole-genome sequencing and tools like antiSMASH. Use homologous recombination with a suicide vector to disrupt a key gene (e.g., the first gene in the operon,
bat1for kalimantacin). This can knock down production of known compounds, simplifying the isolation of novel molecules from the extract [88].
- Procedure: Identify the key biosynthetic gene cluster (BGC) via whole-genome sequencing and tools like antiSMASH. Use homologous recombination with a suicide vector to disrupt a key gene (e.g., the first gene in the operon,
Characterization and Mechanism of Action
Q3: How can we confirm the target of a novel antibiotic that inhibits cell wall synthesis?
- A: A combination of biochemical and biophysical techniques is required.
- Incorporation Assays: First, incorporate radiolabeled precursors (e.g., N-acetylglucosamine, GlcNAc) into major biosynthetic pathways (DNA, RNA, protein, peptidoglycan). Clovibactin specifically blocked GlcNAc incorporation into the cell wall [91].
- Pathway Monitoring: Use reporter systems like
lial-luxinduction. An increase in signal indicates interaction with the lipid II biosynthesis pathway, as was observed with Clovibactin [91]. - Target Binding Validation: Employ Solid-State NMR to study the antibiotic-target complex in a membrane environment. This confirmed Clovibactin binds lipid II's pyrophosphate [90] [91]. Isothermal Titration Calorimetry (ITC) or Surface Plasmon Resonance (SPR) can also provide binding affinity data.
Q4: Our novel antibiotic forms aggregates. Is this part of its mechanism or an artifact?
- A: Aggregation can be a functional mechanism. For Clovibactin, fibril formation was essential.
- Investigation: Use Atomic Force Microscopy (AFM) to visualize structures formed on model bacterial membranes. Clovibactin formed stable, non-toxic fibrils only on membranes containing lipid-anchored pyrophosphates [90] [86].
- Correlation: Correlate fibril formation with antibacterial activity and target sequestration. These fibrils ensured long-lasting, irreversible binding of target precursors, contributing to the lack of resistance [90] [93].
Efficacy and Resistance Studies
Q5: What is the standard to claim "no detectable resistance" against a new antibiotic?
- A: This requires rigorous and prolonged laboratory evolution experiments.
- Protocol: Perform serial passage assays, repeatedly exposing a high inoculum of bacteria (e.g., >10^9 CFU) to sub-lethal concentrations of the antibiotic over many generations (e.g., 20-30 days). For Clovibactin, no resistance was observed in these studies [90] [93]. The resistance frequency should be very low, ideally below <10^-10, which is considered an excellent benchmark [91].
Q6: The antibiotic works in vitro but shows toxicity in mammalian cells. What are the next steps?
- A: This is a common hurdle. Clovibactin showed no cytotoxicity, attributed to its selective action [91] [86].
- Understand Selectivity: Investigate if your compound's target is unique to bacteria. Clovibactin's targets (C55PP, Lipid II, Lipid IIIWTA) are not present in human cells.
- Check Mechanism-Specificity: Confirm if the compound activates non-bacterial pathways. The formation of fibrils specifically on bacterial membranes was a key factor for Clovibactin's low toxicity [86].
- Chemical Modification: If the core scaffold is promising, explore synthetic biology or semi-synthesis to generate analogs with improved therapeutic indices.
The experimental data generated for Clovibactin provides a benchmark for the activity and properties expected from a promising novel antibiotic candidate.
Table 2: Summary of Key Experimental Data for Clovibactin
| Parameter | Result / Value | Experimental Context & Notes |
|---|---|---|
| Minimum Inhibitory Concentration (MIC) | 0.25 - 2 µg/mL | Against a range of Gram-positive pathogens, including MRSA and VRE [88]. |
| Cytotoxicity (Mammalian Cells) | None detected at high concentrations | Indicating selective toxicity for bacterial cells [91] [86]. |
| Resistance Frequency | < 10^-10 | No detectable resistance in serial passage experiments [90] [91]. |
| In Vivo Efficacy | Protected mice from MRSA infection | Successful treatment in a mouse model of S. aureus infection [90] [85]. |
| Key Molecular Targets | C55PP, Lipid II, Lipid IIIWTA | Pyrophosphate moiety of multiple essential cell wall precursors [90] [93]. |
Mechanism of Action: A Visual Guide
Clovibactin exhibits a unique multi-target mechanism that underpins its potency and low resistance profile. The following diagram details the key steps of this mechanism, from binding to bacterial cell death.
FAQs and Troubleshooting Guides
FAQ 1: Why can't I culture the majority of bacteria from my environmental sample in the lab?
This is a common challenge known as the "great plate count anomaly," where the number of colonies on a plate is vastly outnumbered by the microscopic cell count [1]. The primary reason is that standard laboratory conditions fail to replicate essential aspects of the microbes' natural environment [1] [51]. Specific reasons include:
- Incorrect Nutrient Levels: Standard media are often too nutrient-rich, favoring fast-growing bacteria and outcompeting slow-growing or oligotrophic species [51].
- Lack of Essential Growth Factors: Some bacteria depend on specific co-factors, signaling molecules, or nutrients produced by other community members that are absent in pure culture setups [1] [51].
- Inhibitory By-Products of Media Preparation: Autoclaving agar together with phosphate generates hydrogen peroxide, which can inhibit the growth of bacteria that do not produce catalase [94] [51].
- Dependence on Other Microbes: Many bacteria live in symbiotic relationships and require the presence of other bacterial species to grow [1] [6].
- Incorrect Atmospheric or Physical Conditions: The required oxygen levels, temperature, or pH may not be accurately reproduced [95] [96].
FAQ 2: What are the most effective strategies to improve the culturability of uncultured species?
Several innovative strategies have been developed to bring "yet-to-be-cultured" bacteria into cultivation:
- Modify Gelling Agents and Media Preparation: Replace standard agar with gellan gums (Gelrite or Phytagel) and autoclave the gelling agent and phosphate buffer separately. This reduces the formation of hydrogen peroxide and has been shown to significantly increase the number and diversity of cultured bacteria from soil samples [94].
- Use of Simulated Natural Environments: Employ diffusion chambers or membrane-bound devices to incubate bacteria in their natural habitat or a simulated environment. This allows environmental nutrients and growth factors to diffuse in while containing the cells [1] [51].
- Apply Co-culture Techniques: Cultivate your target bacterium together with other bacteria from its environment. Helper strains can provide essential growth factors or signaling molecules [1] [6].
- Incorporate Growth Stimulants: Add resuscitation-promoting factors (Rpf) or sterile soil/water extracts to the culture media to stimulate the growth and resuscitation of dormant cells [6] [51].
- Utilize High-Throughput and Dilution-to-Extinction Methods: These methods use low-nutrient media and single-cell sorting to isolate and grow oligotrophic bacteria that are overwhelmed in standard plates [6].
FAQ 3: How does the choice of growth medium affect the diversity of cultured anaerobic bacteria?
The choice of medium is critical, as different media support the growth of different microbial taxa. A study comparing two media for culturing anaerobic bacteria from human stool found clear differences in community profiles [97].
- Yeast extract cysteine blood agar (HCB) supported the growth of a more diverse microbial community.
- Modified peptone-yeast extract-glucose (MPYG) improved the growth rates of certain specific taxa.
This highlights that there is no single "best" medium. For standard diagnostics, HCB may be preferable, while MPYG might be better for targeting specific bacterial groups [97].
FAQ 4: My bacterial culture is not growing after revival from a frozen or lyophilized state. What should I do?
Some bacterial strains, especially after the stress of cryopreservation or lyophilization, exhibit a prolonged lag phase [96]. Recommended actions include:
- Extended Incubation: Continue to incubate the culture for a longer period, as growth may be delayed [96].
- Check Atmospheric Conditions: Verify that the culture is being incubated in the correct atmosphere (aerobic, microaerophilic, or anaerobic) [95] [96].
- Secondary Transfer: Aseptically transfer a small volume of the primary culture to fresh medium. This can sometimes help resuscitate the culture by diluting accumulated waste products or providing fresh nutrients [96].
Data Presentation: Media and Method Comparison
Table 1: Impact of Gelling Agent and Preparation Method on Bacterial Culturability Data from a study on wheat rhizosphere microbiome cultured on LB medium, showing Colony Forming Units (CFU) from different soils [94].
| Soil Type | Gelling Agent | Phosphate & Gelling Agent Preparation | CFU (x 10ⶠcells mlâ»Â¹) |
|---|---|---|---|
| Clay Loam (Luxor) | Agar | Autoclaved Together | Not Specified (Lowest) |
| Gellan Gum (Phytagel) | Autoclaved Separately | 116.3 ± 27.0 | |
| Sandy (Minya) | Agar | Autoclaved Together | 2.08 ± 0.51 |
| Gellan Gum (Phytagel) | Autoclaved Separately | Not Specified (Highest) | |
| Calcareous (Nubaria) | Agar | Autoclaved Together | Not Specified (Lowest) |
| Gellan Gum (Gelrite/Phytagel) | Autoclaved Separately | Not Specified (Highest) |
Table 2: Relative Performance of Media for Anaerobic Gut Bacteria Cultivation Based on a comparison of culture media for anaerobic bacteria from human stool samples [97].
| Growth Medium | Richness & Evenness | Key Findings |
|---|---|---|
| Yeast extract cysteine blood agar (HCB) | Higher Diversity | Supported a more diverse microbial community; suitable for standard diagnostics. |
| Modified peptone-yeast extract-glucose (MPYG) | Improved Growth for Specific Taxa | Enhanced growth rates of certain bacterial groups; appropriate for targeting specific conditions. |
Detailed Experimental Protocols
Protocol 1: Improved Cultivation Using Alternative Gelling Agents
This protocol is adapted from a study that successfully increased the diversity of cultured bacteria from the wheat rhizosphere [94].
- Prepare Solidifying Agents: Prepare solutions of the chosen gelling agents (e.g., 1.5-2% Agar, Gelrite, or Phytagel) in deionized water.
- Prepare Phosphate Buffer: Prepare a separate phosphate buffer solution (e.g., 10 mM, pH 7.0).
- Separate Sterilization: Autoclave the gelling agent solution and the phosphate buffer solution separately to prevent the formation of hydrogen peroxide.
- Prepare Nutrient Base: Prepare and autoclave the nutrient base of your chosen medium (e.g., LB or Jensen medium) without phosphate.
- Mix Medium: After all solutions have cooled sufficiently (to ~50-60°C to prevent solidification), mix the nutrient base, phosphate buffer, and gelling agent together under sterile conditions.
- Pour Plates: Pour the mixed medium into sterile petri dishes.
- Inoculate and Incubate: Inoculate the plates with your sample using standard streaking techniques and incubate at the appropriate temperature.
Protocol 2: Dilution-to-Extinction Culturing for Oligotrophic Bacteria
This high-throughput method is useful for isolating slow-growing bacteria from aquatic and terrestrial environments [6].
- Sample Inoculum: Prepare a dilute cell suspension from your environmental sample (e.g., soil, water) in a sterile saline solution.
- Prepare Low-Nutrient Media: Prepare a large volume of a low-nutrient, defined liquid medium that mimics the ionic composition of the sample's environment.
- Dilution Series: Using a sterile dispenser, aliquot the dilute cell suspension into multi-well plates (e.g., 96-well plates) containing the low-nutrient medium. The goal is to statistically inoculate many wells with a single bacterial cell.
- Long-Term Incubation: Seal the plates to prevent evaporation and incubate them at the in situ temperature for extended periods (weeks to months).
- Monitor Growth: Monitor the wells for turbidity, which indicates bacterial growth.
- Subculture: Once growth is detected, subculture from positive wells onto solid media to obtain pure isolates.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Microbial Cultivation Experiments
| Item | Function/Application |
|---|---|
| Gellan Gums (Gelrite, Phytagel) | Alternative gelling agents to agar; produce less hydrogen peroxide during autoclaving, improving recovery of fastidious bacteria [94]. |
| Resuscitation-Promoting Factor (Rpf) | A bacterial growth factor from Micrococcus luteus that stimulates the resuscitation and growth of dormant cells in environmental samples [6]. |
| Diffusion Chambers | Semi-permeable membranes used to incubate bacteria in their natural environment, allowing diffusion of nutrients and growth factors from the surroundings [1] [51]. |
| Selective Media (e.g., MacConkey Agar) | Contains inhibitors (e.g., bile salts, crystal violet) to suppress unwanted bacteria and differential components (e.g., lactose) to identify specific groups based on colony color [95]. |
| Anaerobic Chamber/Glove Box | Creates an oxygen-free environment for the cultivation, manipulation, and storage of strict anaerobic bacteria [95]. |
Workflow Visualization: Troubleshooting Unculturable Bacteria
Frequently Asked Questions
Q1: Why is my dilution-to-extinction cultivation yielding no growth for target oligotrophs? This typically occurs due to improper media composition or cell density. For freshwater oligotrophs, use defined artificial media mimicking natural conditions with organic compounds in µM concentrations (1.1-1.3 mg DOC per liter) [10]. Ensure your inoculum contains approximately one cell per well in 96-deep-well plates, and incubate for 6-8 weeks at environmentally relevant temperatures (e.g., 16°C for lake microbes). The viability rate is typically around 12.6%, so sufficient replication is crucial [10].
Q2: How can I determine if my uncultured bacterium requires specific growth factors from helper organisms? When pure culture attempts consistently fail despite optimizing physical parameters, coculture dependence should be investigated. Evidence includes: growth only in spent media from other bacteria [98], or formation of microcolonies only in close proximity to helper strains in diffusion chambers [1]. Molecular analysis can identify specific dependencies; for instance, some strains require siderophores like enterobactin produced by helper organisms [98].
Q3: What metabolic tracing approach should I use to track nutrient utilization in my newly cultured isolate? Use stable isotope tracers (e.g., 13C-glucose) introduced via incubation in cell culture media [99]. Select atoms that won't be lost as CO2 before reaching your metabolites of interest. For detecting pathway activities rather than just rates, track labeling patterns in downstream metabolites using mass spectrometry. Exposure time should match process kinetics - detecting labels in synthesized proteins requires longer experiments than measuring glycolytic lactate production [99].
Q4: Why does my topological pathway analysis consistently highlight the same ubiquitous metabolites? This over-emphasis on hub compounds (e.g., ATP, glutamate) is a known limitation of betweenness centrality calculations in connected metabolic networks [100]. Apply a hub penalization scheme to diminish the disproportionate influence of these compounds. Alternatively, analyze pathways both connected and disconnected from the global network, as disconnected analysis prevents hub over-emphasis while connected analysis provides physiological context [100].
Q5: How can I validate whether genomic predictions of metabolic capabilities match actual function in my new isolate? Combine metabolic tracing with elementary mode analysis. Grow cells with 13C-labeled substrates (e.g., glucose or glutamine) and track incorporation into downstream metabolites. Compare the actual labeling patterns with those predicted by elementary modes - the minimal sets of enzymes that can support steady-state operation [101]. Discrepancies indicate missing regulatory understanding or incorrect pathway annotations.
Troubleshooting Guides
Common Cultivation Challenges
Table 1: Troubleshooting Microbial Cultivation Issues
| Problem | Potential Causes | Solutions | Supporting Evidence |
|---|---|---|---|
| No growth in dilution-to-extinction | Media too rich, inhibiting oligotrophs | Use defined low-nutrient media (1-2 mg DOC/L); avoid carbon sources >10μM [10] | Lake isolates achieved 40% viability with optimized media [10] |
| Growth only in diffusion chambers | Missing soluble factors from environment | Recreate environment via cell-free environmental extracts; use semi-permeable membranes [1] [98] | Recovery increased from 0.05% to 40% using environmental nutrients [98] |
| Growth dependent on other species | Coculture requirements for growth factors | Identify helper strains; supplement with spent media; add siderophores or vitamins [98] | Enterobactin identified as essential growth factor for Maribacter polysiphoniae [98] |
| Initial growth ceases upon transfer | Accumulation of toxic metabolites | Add catalase to media; use continuous culture; reduce cell density [10] | Catalase included in successful freshwater media formulations [10] |
Metabolic Analysis Challenges
Table 2: Troubleshooting Metabolic Pathway Analysis
| Problem | Potential Causes | Solutions | Supporting Evidence |
|---|---|---|---|
| Pathway analysis identifies unrealistic pathways | Inclusion of non-host reactions | Use organism-specific pathway definitions; filter non-native enzymatic reactions [100] | Generic vs. human-only pathways show significant outcome differences [100] |
| Metabolomics shows changes but no mechanism | Static snapshots without dynamics | Implement metabolic tracing with 13C-labeling; measure flux over time [99] | Tracing revealed gut microbiome processes in NAD synthesis [99] |
| Elementary modes don't match experimental data | Missing regulatory constraints | Incorporate known allosteric regulation; measure enzyme activities [101] | Elementary modes provide structure but require biological validation [101] |
| Central metabolites dominate network analysis | Over-weighting of hub compounds | Apply betweenness centrality with penalization; analyze subnetworks [100] | Hub penalization schemes prevent over-emphasis of ubiquitous metabolites [100] |
Experimental Protocols
High-Throughput Dilution-to-Extinction Cultivation
Purpose: Isolate previously unculturable oligotrophic bacteria from environmental samples.
Materials:
- Sterile 96-deep-well plates
- Defined low-nutrient media (e.g., med2/med3 with 1.1-1.3 mg DOC/L) [10]
- Environmental sample (water, soil extract)
- Sterile dilution blanks
- Incubation system (16°C for freshwater isolates)
Procedure:
- Prepare three defined media types: carbohydrate-based (med2), organic acid-based (med3), and C1 compound-based (MM-med) for metabolic diversity [10].
- Serially dilute environmental sample to approximately 1 cell/well in 96-deep-well plates.
- Incubate at in situ temperature (16°C for lakes) for 6-8 weeks without disturbance.
- Screen for growth visually or via fluorescence microscopy.
- Transfer positive wells to fresh media; confirm purity by 16S rRNA sequencing.
- Characterize growth rates in multiple media types to classify as oligo-, meso-, or copiotrophs.
Validation: Successful isolation should capture taxa representing up to 72% of genera detected in original metagenomic samples [10].
Stable Isotope Metabolic Tracing
Purpose: Track nutrient utilization through metabolic pathways in new isolates.
Materials:
- 13C-labeled substrates (e.g., glucose, glutamine)
- Mass spectrometry system
- Defined growth media
- Quenching solution (cold methanol)
- Extraction solvents
Procedure:
- Grow cells to mid-exponential phase in standard media.
- Transfer to media containing 13C-labeled tracer substrates at same concentration.
- Collect samples at multiple time points (seconds to hours depending on process kinetics).
- Quench metabolism immediately with cold methanol.
- Extract metabolites using appropriate solvents.
- Analyze label incorporation patterns via LC-MS or GC-MS.
- Calculate isotopic enrichment and map to metabolic pathways.
Interpretation: Compare labeling patterns to predictions from genomic annotations. Faster-than-expected labeling may indicate preferred nutrient usage [99].
Experimental Workflows
Cultivation Workflow for Unculturable Species
Metabolic Pathway Analysis Workflow
Research Reagent Solutions
Table 3: Essential Research Reagents for Phenotypic Characterization
| Reagent/Category | Specific Examples | Function/Application | Key Considerations |
|---|---|---|---|
| Defined Media Components | med2, med3, MM-med [10] | Cultivation of oligotrophic freshwater bacteria | Low carbon (1.1-1.3 mg DOC/L); includes carbohydrates, organic acids, vitamins |
| Isotope Tracers | 13C-glucose, 13C-glutamine [99] | Metabolic flux analysis; pathway activity measurement | Choose atoms that persist through pathways; concentration matching endogenous levels |
| Pathway Analysis Tools | MetaboAnalyst, KEGG [100] | Functional interpretation of metabolomics data | Use organism-specific pathways; apply hub penalization for connected networks [100] |
| Cultivation Equipment | Diffusion chambers [98] | Growth in simulated natural conditions | Semi-permeable membranes allow nutrient exchange while containing cells |
| Metabolic Inhibitors | Succinylacetone, Ivosidenib [102] | Targeted pathway disruption; therapeutic testing | Succinylacetone inhibits heme synthesis; Ivosidenib arrests TCA cycle [102] |
Conclusion
The paradigm that the majority of microorganisms are unculturable is being rapidly overturned by innovative strategies that more faithfully replicate natural environments and leverage genomic insights. The successful cultivation of novel taxa, including abundant yet previously elusive freshwater oligotrophs and human gut microbes, is unlocking a new era of discovery. This progress is not merely academic; it directly fuels the pipeline for novel therapeutics, as exemplified by the discovery of the unique antibiotic clovibactin. The future of microbial cultivation lies in the continued integration of high-throughput technologies, refined environmental simulation, and multi-omics data, promising to transform these newly cultured isolates into model organisms for ecological study and powerful engines for biomedical innovation.
References
- 1. Growing Unculturable Bacteria - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3416243/]
- 2. The Great Plate Count Anomaly - by Allan Konopka [https://thinkmicrobe.substack.com/p/the-great-plate-count-anomaly]
- 3. Uncultured Microorganisms and Their Functions in the ... [https://www.mdpi.com/2304-8158/12/14/2691]
- 4. Development of a novel cultivation technique for ... [https://www.nature.com/articles/s41598-019-43182-x]
- 5. High-Throughput Methods for Culturing Microorganisms in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC124033/]
- 6. The Uncultured Microorganisms: Novel Technologies and ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.756287/full]
- 7. The great screen anomaly—a new frontier in product ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3264863/]
- 8. Effects of Mass Culture Technique on Microbial Yield and ... [https://digitalcommons.pcom.edu/research_day/research_day_GA_2018/researchGA2018/18/]
- 9. Exploring the antibiotic potential of cultured 'unculturable ... [https://www.sciencedirect.com/science/article/abs/pii/S0966842X2300330X]
- 10. Bringing the uncultivated microbial majority of freshwater ... [https://www.nature.com/articles/s41467-025-63266-9]
- 11. Frontiers | Editorial: Exploring the diversity, ecological significance, and... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1604849/full]
- 12. New Strategies for Cultivation and Detection of Previously ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC492380/]
- 13. Opportunities and challenges of using metagenomic data to bring... [https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-022-01272-5]
- 14. The Uncultured Microorganisms: Novel Technologies and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8652222/]
- 15. Culturing uncultivated bacteria | Nature Methods [https://www.nature.com/articles/s41592-019-0634-1]
- 16. Faster and more affordable sequencing with the HiFi ... [https://www.pacb.com/blog/high-throughput-long-read-protocols/]
- 17. Strategies for Natural Products Discovery from Uncultured ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8156983/]
- 18. Towards the sustainable discovery and development of ... [https://www.nature.com/articles/s41570-021-00313-1]
- 19. Few antibiotics under development – How did we end up here? [https://www.reactgroup.org/toolbox/understand/how-did-we-end-up-here/few-antibiotics-under-development/]
- 20. Bacterial Natural Product Drug Discovery for New Antibiotics [https://pmc.ncbi.nlm.nih.gov/articles/PMC8300778/]
- 21. Current economic and regulatory challenges in developing ... [https://www.nature.com/articles/s44259-025-00123-1]
- 22. Challenges of Antibacterial Discovery - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3021209/]
- 23. Mining Microbial Dark Matter: Advanced Cultivation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12566907/]
- 24. Isolation of Uncultured Bacteria from Antarctica Using Long ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2017.01346/full]
- 25. protocols.io/view/high-throughput- cultivation -and- dilution - to -extinc-du... [https://www.protocols.io/view/high-throughput-cultivation-and-dilution-to-extinc-du336yqn.html]
- 26. Dilution-to-extinction - (General Biology I) [https://fiveable.me/key-terms/college-bio/dilution-to-extinction]
- 27. Evaluating agar-plating and dilution-to-extinction isolation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11878766/]
- 28. Culturing of “Unculturable†Subsurface Microbes: Natural ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2020.610001/full]
- 29. High molecular weight dissolved organic matter enrichment selects for... [https://www.nature.com/articles/ismej201568?error=cookies_not_supported&code=4efe07d2-d646-47af-9f66-6fa8c19a27ac]
- 30. High-throughput cultivation based on dilution - to - extinction with... [https://link.springer.com/article/10.1007/s12275-020-0452-2]
- 31. Microencapsulation and in situ incubation methodology for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9441948/]
- 32. How to troubleshoot bioreactor contamination [https://infors-ht.com/en/blog/how-to-troubleshoot-bioreactor-contamination]
- 33. Best Practices For Designing Microbiology Experiments [https://www.outsourcedpharma.com/doc/best-practices-for-designing-microbiology-experiments-0001]
- 34. Formation of a constructed microbial community in a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11019790/]
- 35. Strategies and tools to construct stable and efficient artificial ... [https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-024-02594-2]
- 36. Computer-guided design of optimal microbial consortia for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5959721/]
- 37. Engineering Microbial Consortia as Living Materials [https://pmc.ncbi.nlm.nih.gov/articles/PMC11421429/]
- 38. Cell Culture Contamination Troubleshooting [https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/cell-culture-and-cell-culture-analysis/mammalian-cell-culture/cell-culture-troubleshooting-contamination?srsltid=AfmBOorpSAVnIKN2dr-cKhKtJCDJeev1YA2cP2-UBALW8CD-DkPyIZrS]
- 39. Troubleshooting Cell Culture Contamination: A Practical ... [https://www.yeasenbio.com/blogs/cell/troubleshooting-cell-culture-contamination-a-practical-solution-guide]
- 40. Troubleshooting Common Cell Culture Contamination Issues [https://cellculturecompany.com/troubleshooting-common-cell-culture-contamination-issues/]
- 41. Insights into the Cultured Bacterial Fraction of Corals - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8269258/]
- 42. Prediction of Bacterial Growth on Different Culture Media ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12495271/]
- 43. Advances in the isolation, cultivation, and identification of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11145792/]
- 44. Optimized high-throughput whole-genome sequencing ... [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-025-01512-x]
- 45. Implementation of a high-throughput whole genome ... [https://pubmed.ncbi.nlm.nih.gov/38451098/]
- 46. Optimizing High-Throughput Screening Workflows in ... [https://kanboapp.com/en/optimizing-high-throughput-screening-workflows-in-scientific-research-a-guide-to-improved-efficiency-and-data-integrity/]
- 47. High-throughput workflow for cultivation and ... [https://www.nature.com/articles/s41598-025-93180-5]
- 48. High-Throughput Sequencing: Definition, Technology, ... [https://www.cd-genomics.com/resource-comprehensive-overview-high-throughput-sequencing.html]
- 49. High-throughput computing and workflows - Docs CSC [https://docs.csc.fi/computing/running/throughput/]
- 50. Common Workflow Problems: Identify and Solve Them [https://hyscaler.com/insights/common-workflow-problems-solve-them/]
- 51. Culturing the Unculturable: Working with Difficult Bacteria [https://bitesizebio.com/44193/culturing-the-unculturable-working-with-difficult-bacteria/]
- 52. A simple nutrient-dependence mechanism for predicting the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4500256/]
- 53. Community Interaction Co-limitation: Nutrient Limitation in ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2022.846890/full]
- 54. Nutrient Stoichiometry Shapes Microbial ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2017.00949/full]
- 55. Nutrient-Limited Operational Strategies for the Microbial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9695796/]
- 56. Double emulsions as a high-throughput enrichment and isolation... [https://www.nature.com/articles/s43705-023-00241-9?error=cookies_not_supported]
- 57. Agar medium for microbial growth [https://www.sigmaaldrich.com/TW/zh/search/agar-medium-for-microbial-growth?focus=sitecontent&page=1&perpage=30&sort=relevance&term=agar%20medium%20for%20microbial%20growth&type=site_content&srsltid=AfmBOooLmCKqZvXgKM5KyRQmw2oYzhQTK8DJv0HRCRtJaOSF8JkARTkC]
- 58. Building microbial communities to improve antimicrobial ... [https://www.nature.com/articles/s44259-025-00115-1]
- 59. The evolving copiotrophic/oligotrophic dichotomy: From Winogradsky to... [https://pubmed.ncbi.nlm.nih.gov/36856667/]
- 60. Interaction between bacterial endophytes and host plants - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9890182/]
- 61. Perspectives and potential applications of endophytic ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.985429/full]
- 62. Tiny magnetic capsules help grow unculturable microbes [https://cen.acs.org/biological-chemistry/biotechnology/Tiny-magnetic-capsules-help-grow/103/web/2025/11]
- 63. Microbe Products for Research [https://www.atcc.org/microbe-products]
- 64. Culturing of 'unculturable' human microbiota reveals novel ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4890681/]
- 65. Oxygen Restriction Generates Difficult-to-Culture P. ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2019.01992/full]
- 66. Biological pH buffers in IVF: help or hindrance to success - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3170115/]
- 67. Dual- Temperature of Cellular Products... Microbiological Control [https://pubmed.ncbi.nlm.nih.gov/37764194/]
- 68. 17.2: Temperature - Biology LibreTexts [https://bio.libretexts.org/Learning_Objects/Laboratory_Experiments/Microbiology_Labs/Microbiology_Labs_II/17%3A_Use_of_Physical_Agents_to_Control_of_Microorganisms/17.02%3A_Temperature]
- 69. Noninvasive Methods for the Investigation of Organisms at ... [https://www.sciencedirect.com/science/article/abs/pii/S0065216402510054]
- 70. Multi-omic approaches for host-microbiome data integration [https://pmc.ncbi.nlm.nih.gov/articles/PMC10766395/]
- 71. Multi-omic integration of microbiome data for identifying ... [https://www.nature.com/articles/s41467-024-46888-3]
- 72. Unexplored microbial diversity from 2500 food ... [https://www.sciencedirect.com/science/article/pii/S009286742400833X]
- 73. Integration of metagenome-assembled genomes with ... [https://www.nature.com/articles/s41467-025-64950-6]
- 74. Validation Error Explanations for Genomes - NCBI - NIH [https://www.ncbi.nlm.nih.gov/genbank/genome_validation]
- 75. Unraveling metagenomics through long-read sequencing [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-024-04917-1]
- 76. Opportunities and challenges in long-read sequencing data ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-020-1935-5]
- 77. Sequencing 101: long-read sequencing [https://www.pacb.com/blog/long-read-sequencing/]
- 78. Genome-resolved long-read sequencing expands known ... [https://www.nature.com/articles/s41564-025-02062-z]
- 79. How HiFi sequencing achieves scalable microbial ... [https://www.pacb.com/blog/how-hifi-sequencing-achieves-scalable-microbial-genomics-without-compromise/]
- 80. Software and bioinformatics powering long-read sequencing [https://www.pacb.com/blog/sequencing-101-software-and-bioinformatics-powering-long-read-sequencing/]
- 81. Genome-resolved long-read sequencing expands known ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12313526/]
- 82. Closing the genome of unculturable cable bacteria using a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10926707/]
- 83. Exploring high-quality microbial genomes by assembling ... [https://www.nature.com/articles/s41467-024-49060-z]
- 84. Editorial: Long-read sequencing—Pitfalls, benefits and ... [https://www.frontiersin.org/articles/10.3389/fgene.2022.1114542/full]
- 85. Powerful new antibiotic isolated from microbial 'dark matter' [https://www.ddw-online.com/powerful-new-antibiotic-isolated-from-microbial-dark-matter-25500-202308/]
- 86. New antibiotic from microbial 'dark matter' could be ... [https://www.uu.nl/en/news/new-antibiotic-from-microbial-dark-matter-could-be-powerful-weapon-against-superbugs]
- 87. Microbial 'dark matter' yields new antibiotic [https://www.nature.com/articles/d41573-023-00156-z]
- 88. A new antibiotic from an uncultured bacterium binds to an ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10245560/]
- 89. Clovibactin and Staphylococcus aureus: a new weapon ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11565595/]
- 90. An antibiotic from an uncultured bacterium binds to ... [https://www.sciencedirect.com/science/article/pii/S009286742300853X]
- 91. Instruct-NL Develop Clovibactin: The Potential New ... [https://instruct-eric.org/scientifichighlights/instruct-nl-develop-clovibactin-the-potential-new-antibiotic-with-low-resistance/]
- 92. Clovibactin - a game-changing novel antibiotic with no ... [https://www.news-medical.net/news/20230906/Clovibactin-a-game-changing-novel-antibiotic-with-no-resistance.aspx]
- 93. New Antibiotic, Clovibactin, Kills Bacteria Without ... [https://www.genengnews.com/topics/infectious-diseases/new-antibiotic-clovibactin-kills-bacteria-without-developing-resistance/]
- 94. Comparative Analysis of the Cultured and Total Bacterial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8528112/]
- 95. How to culture bacteria - INTEGRA Biosciences [https://www.integra-biosciences.com/united-states/en/blog/article/how-culture-bacteria]
- 96. Bacteriology Culture Guide [https://www.atcc.org/resources/culture-guides/bacteriology-culture-guide]
- 97. Comparison of the diversity of anaerobic-cultured gut ... [https://www.sciencedirect.com/science/article/pii/S0167701224001003]
- 98. Unculturable Bacteria and Their Significance in Contemporary ... [https://microbewiki.kenyon.edu/index.php/Unculturable_Bacteria_and_Their_Significance_in_Contemporary_Medicine]
- 99. A Beginner's Guide to Metabolic Tracing [https://bitesizebio.com/81841/metabolic-tracing-guide/]
- 100. Metabolic Pathway Analysis: Advantages and Pitfalls for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9953275/]
- 101. Elementary mode analysis: a useful metabolic pathway ... [https://pubmed.ncbi.nlm.nih.gov/19015845/]
- 102. Metabolic pathway [https://en.wikipedia.org/wiki/Metabolic_pathway]